Retrocausal Teleportation Protocol
Time travel has long captured the human imagination, relegated to the realm of science fiction and fantasy. However, recent developments in quantum physics suggest that, at least at the quantum level, retrocausal effects—where future events influence the past—may be not only possible but experimentally testable. The retrocausal teleportation protocol, proposed by researchers from institutions including Hitachi Cambridge Laboratory, University of Cambridge, and the University of Maryland, represents a groundbreaking approach to exploring these temporal paradoxes through the lens of quantum mechanics.
While classical physics presents a deterministic universe where cause must precede effect, quantum mechanics and relativity theory paint a more nuanced picture. There are already well-known examples from relativity theory like wormholes, which are valid solutions of Einstein's Field Equations, and similarly in quantum mechanics the non-classical state of quantum entanglement—the "spooky action at a distance" that troubled Einstein—which demonstrates that quantum systems can maintain instantaneous correlations across space and, potentially, time.
Perhaps most intriguingly, the protocol suggests that quantum entanglement can be used to effectively send information about optimal measurement settings "back in time"—information that would normally only be available after an experiment is complete. This capability, while probabilistic in nature, could revolutionize quantum computing and measurement techniques. Recent advances in multipartite hybrid entanglement even suggest these effects might be achievable in real-world conditions, despite environmental noise and interference.
This article explores the theoretical foundations, experimental proposals, and potential applications of the retrocausal teleportation protocol. From its origins in quantum mechanics and relativity theory to its implications for our understanding of causality and the nature of time itself, we examine how this cutting-edge research challenges our classical intuitions while opening new possibilities for quantum technology. As we delve into these concepts, we'll see how the seemingly fantastic notion of time travel finds a subtle but profound expression in the quantum realm, potentially revolutionizing our approach to quantum computation and measurement while deepening our understanding of the universe's temporal fabric.
Introduction
Classical Newtonian physics painted a picture of the world that was predictable, deterministic, and some might even say common sensical. Physical systems and particle-to-particle interactions can be explained by force, mass (F = ma) and Newton's laws of motion. Also, a seemingly common sense notion from classical mechanics is that there are very definite cause-and-effect relationships, with things that happen in the past determining present conditions and never the other way around. In classical physics this would make it possible, at least theoretically, to completely describe the past state and future trajectory of a system with absolute precision by extrapolating from its present configuration. However, with relativity and quantum mechanics, what are called non-classical physics, the behavior of nature becomes a little more—let’s say— “non-linear”, and even within the standard model some classical notions associated with Newtonian mechanics become obsolete.
One such situation is what are called closed time-like curves (CTCs), which are essentially “loops” in spacetime that can naturally emerge from Einstein field equations (EFE) under certain circumstances—for instance CTCs are naturally emergent in Gödel solution of EFE—and describe a trajectory (called a world line in relativity) that returns to the same point in space and time, effectively describing a spacetime geometry permitting time travel, or at the very least losing the causal distinction of “before” and “after” because, as Einstein described it: “…the distinction ‘earlier-later’ is abandoned for world-points which lie far apart in a cosmological sense, and those paradoxes, regarding the direction of the causal connection, arise, of which Mr. Gödel has spoken” [1].
So, we see that closed time-like curves arise with relative ease in relativity and are predicted in natural systems, for example within the ergosphere of black holes. These causally ambiguous loops also arise within quantum physics because the quantum vacuum is an entangled state, with both non-classical correlations in space-like and time-like regions. That means non-local connections between causally separated regions in space and time, i.e., closed time-like curves. This gives rise to effects like Unruh thermalization and emission of photons from the vacuum—recently observed and measured—which occurs because the vacuum state of the electromagnetic field has intrinsic quantum entanglement between past and future states [2, 3, 4]. This raises the question, can entanglement be used to influence “past” events?
The fact that CTCs are found in both relativity and quantum mechanics has a potentially straight-forward answer, which can speak to the physical relevance of closed-timelike loops in spacetime. The solution comes when it is realized that quantum entanglement—sometimes referred to as Bell pairs or Einstein-Podolsky-Rosen correlations (EPR)—is multiply connected spacetime geometry, also known as Einstein-Rosen bridges (ER) such that ER = EPR [5] (a resolution to the EPR paradox first proposed by Einstein, Podolsky, and Rosen). So, both relativity and QM say that time travel is possible, with some observable effects being the result of non-local connections or spacetime entanglement “loops”, and aspects as fundamental as time symmetry seeming to require retrocausality [6, 7].
Interesting results begin to emerge when considering quantum field theory in curved spacetimes (QFT in CST)—we will just focus on some specifics here and not the more general ramifications of a Unified Field Theory— because we can analyze the behavior of quantum systems, like qubits, in the presence of closed time-like curves, for example modeling the nonlinearities that arise from a qubit interacting with an older version of itself in a CTC [8]. The study of these systems provides valuable insight into nonlinearities and the emergence of causal structures in quantum mechanics—essential for any formulation of a quantum theory of gravity.
Significantly, they have implications for quantum theories of time travel. In quantum physics there is an experimental program generally referred to as quantum teleportation (QT) that enables the transfer of quantum information states via a classical channel by leveraging the strong correlation between quantum entangled systems (QT protocols have also been utilized to transfer energy and may soon verify the ER = EPR conjecture). Standard quantum teleportation relies on quantum entanglement and classical communication to transfer the state of a particle from one location to another without directly transferring the particle itself (the classical communication channel ensures that signals are not transferred instantaneously or faster than light).
Understanding Quantum Teleportation
Before we explore the proposed experiment to send a quantum state “back” in time, a general understanding of the QT protocol must be established. Quantum teleportation transfers a quantum state between two parties (Alice and Bob) using entanglement and classical communication (Figure 1). The key steps are:
Entanglement Creation:
Alice and Bob share an entangled pair of qubits.
State to Teleport:
Alice has an additional qubit with the quantum state ∣ψ⟩| she wants to teleport.
Bell-State Measurement:
Alice performs a Bell-state measurement on her qubit (to be teleported) and her half of the entangled pair, collapsing the system into one of four possible entangled states.
Classical Communication:
Alice sends the measurement result (two classical bits) to Bob.
State Reconstruction:
Bob applies a corresponding quantum operation (e.g., X, Z gates) to his qubit based on Alice’s result, recreating ∣ψ⟩| perfectly.
Note: The original state ∣ψ⟩| is destroyed during the process, preserving the no-cloning theorem.
Figure 1. Illustration of the QT protocol. The protocol consists of four steps. In the first step, the entangled qubit is shared between Alice and Bob. With local operations, Alice entangles the target qubit with the entangled pair (step 2). Then, Alice measures both of her qubits (step 3) and tells Bob the result of the measurement via classical communication. He can then correct his qubit (step 4) and Alice’s state is teleported to Bob (Step 5). Image and image description from Fitzgerald 2024 https://www.researchgate.net/publication/379970865_A_Christmas_story_about_quantum_teleportation
The State of the Art
Advancements in QT protocols have significantly benefited from developments in entanglement swapping and the efficiency of Bell state measurements (BSMs), the latter being named after the famous Bell inequality formulations that define the non-classical nature of certain measurement outcomes with quantum systems, like photonic qubits (entangled photons used to create quantum bits). These innovations are pivotal for enhancing the reliability and scalability of quantum communication networks, which gives direct technological applicability of QT techniques. For instance, entanglement swapping enhancements have including extended entanglement distribution, in which entanglement swapping enables the entanglement of two particles that have never interacted directly, facilitating the extension of entangled links across distant nodes in a quantum network (this technique is similar to entanglement harvesting protocols, which we have recently discussed in recent articles). This principle is fundamental for the development of quantum repeaters, which are essential for long-distance quantum communication. As well improvements in BSMs have been realized with techniques such as boosted quantum teleportation [9].
Combining entanglement swapping with improved BSM efficiency is crucial for the development of quantum repeaters. These devices facilitate the distribution of entanglement over long distances by segmenting the communication channel into shorter links and swapping entanglement between them [10]. As well, these advancements should facilitate the design of hybrid quantum networks, since advances in entanglement swapping protocols have enabled the interconnection of heterogeneous quantum systems, such as linking discrete-variable and continuous-variable nodes, essentially utilizing both the particle (discrete) and wave (continuous) nature of light [11]. This capability is vital for the realization of versatile and robust quantum networks.
QT is not only a key enabler for the development of quantum computing systems and networks but also a cornerstone for secure, tamper-proof communication protocols. These applications hold promises for advancing technology in areas such as cryptography, distributed computing, and global connectivity. Recent studies have shown that these possibilities are nearing experimental realization, the second step in full implementation of quantum non-locality in technology.
For those with an eye to the future, there is an exciting new class of teleportation protocols that may enable remarkable experimental tests of quantum theory, for example to test retrocausality in quantum mechanics—the ability to perform operations to change the past state of a quantum system, like a qubit—for example by performing post-selected teleportation (P-CTCs) [12]. If entanglement is a kind of spatial nonlocality, retrocausal action is a kind of temporal nonlocality, and should be just as realizable as entanglement of spatially separated quanta. This would mean that just as entanglement is utilized in quantum teleportation protocols for quantum networks and computing, these may be vastly advanced with retrocausal entanglement protocols that can improve quantum robustness by leaps and bounds. As well as revealing fundamental properties of our reality, like retrocausal or trans-temporal interactions (nonlocality in space and time), in which future or present states can affect—or be the cause of in a retroactive way—past states.
1.1 The Quantum Mechanics of Time Travel
Researchers have found that the quantum mechanics of closed timelike curves does allow for quantum time travel, which differs from the classical conception of time travel in a few ways, most namely of which is that proposed protocols do not enable paradoxical causal violations and primarily have implications for enhancing quantum measurements—what is called quantum metrology, which are techniques that can leverage certain effects to overcome limits on conventional observation and measurement—and potentially boosting the power of computation.
The interesting thing about microscopic or quantum phenomena is that they can have retrocausal mechanics without violating relativity theory or the strong principle of causality: that a cause must always precede its effects. In relativity, the strong principle can be restated as ‘no information can be sent faster than the speed of light’, which can be called the weak principle of causality. A clear example of how quantum mechanics can be retrocausal without apparent violation of the strong or weak principles of causality is given in David Pegg’s review Retrocausality and Quantum Measurement [13]. Classical electromagnetism (EM) involves quadratic equations that when solved give two answers, which essentially amount to a solution in which an oscillating charge (an emitter) generates EM radiation that goes “forward in time” (what for historical reasons was called “retarded potentials”) and simultaneously an absorber that has sent advanced radiation before the emitter has sent the forward potentials and the energy that is dissipated by the emitter is due to the radiative reaction force of the absorber, such that preceding event (emitted radiation and loss of energy by the charged oscillator) is caused by the future event (absorption of radiation).
Since the advanced radiation involves advanced potentials that act retrocausally on the emitter, this is omitted in the classical interpretation because it would seem to violate the strong principle of causality. However, two brilliant physicists John Wheeler and Richard Feynman re-evaluated the theory of EM and constructed a formulation that retains both the retarded and advanced potentials: what is called Wheeler-Feynman absorber theory of radiation. This theory forms the basis of Cramer’s Transactional Interpretation of quantum mechanics, and although absorber theory contravenes the strong principle of causality, in a universe with perfect absorbing properties it does not violate the weak principle (no macroscopic messages can be sent back in time between two observers) and it predicts precisely the same experimentally verifiable results as the conventional, temporally-unidirectional theory of electrodynamics.
There are other examples in which a retrocausal explanation seems to be more precise to the actual mechanics involved, such as the retrodiction problem in quantum mechanics— Quantum retrodiction involves finding the probabilities for various preparation events given a measurement event—in which backward time evolution of a BSM on an unknown quantum state to influence the preparation of the state before it is sent is shown to be the easiest explanation [14].
The quantum picture that is retrocausal in the sense of violating the strong but not the weak principle of causality can be extended beyond the use of a state evolving backwards in time for retrodictive purposes… we use a retrocausal quantum picture to examine the possibility of preparing a qubit state evolving forwards in time in the normal manner, then sending this state into the past where it appears as an identical forward evolving qubit state that can be measured in the normal manner or used for other purposes. We see that this can only be done on a probabilistic basis that does not violate weak causality, but it is still useful for the main application… that of measuring a quantum optical qubit state before it is prepared.
In the example given by Pegg, CTCs in quantum mechanics could enable instantaneous quantum communication without violating the weak principle of causality (no FTL signaling). While the existence of closed timelike curves is hypothetical, they can be simulated probabilistically by QT circuits.
Can quantum mechanics allow us to effectively send information back in time? Research is needed to answer this question, and progress has recently been made with a new study by researchers from Hitachi Cambridge Laboratory, University of Cambridge, Paul Scherrer Institute, ETH Zürich, and the University of Maryland. The researchers have devised an experiment to test the question of quantum non-locality in time with a novel twist on the long-standing hypothetical method of P-CTCs, quantum retrodiction, and instantaneous quantum computation. In their paper "Nonclassical Advantage in Metrology Established via Quantum Simulations of Hypothetical Closed Timelike Curves," the researchers describe a thought experiment where quantum entanglement is used to simulate sending information backwards in time through a CTC [15], via the basic procedures of QT, but with the inclusion of a particle in a specially prepared quantum state that is sent "back in time" via interaction of entangled EPR pairs (a kind of entangled quantum circuit) to change a particle's state in the past; a retrocausal mechanism. While actual time travel remains in the realm of science fiction, the researchers show how quantum circuits can probabilistically simulate CTCs in a way that provides a practical advantage for quantum measurements. The protocol is outlined as follows (Figure 2):
Figure 2. Schematic outline of the steps involved in sending a quantum state “back” in time for a post-selected teleportation measurement or P-CTC. Image from Quantum time travel: The experiment to 'send a particle into the past', By Miriam Frankel, for NewScientist, May 2024.
It can be seen that the basic premise follows elements of the conventional teleportation protocol, however instead of transferring the quantum state of particle A to particle D, particle D is prepared in the ideal state that the experimenter wants particle A to be in, which upon interaction of particle A’s entangled particle pair, B, there is a small probability that the Bell state measurement will propagate “backwards” in time to project onto particle A, thus retrocausally changing particle A’s quantum state to the desired spin. Since this can’t be utilized to send messages back in time, it does not violate the weak principle of causality, and hence is fully compatible with relativity, which in any case has solutions that are CTCs. Do CTCs like the ‘loop the loop’ of retrocausal teleportation offer empirical verification of the CTCs in relativity? So that they should not be discarded like the advanced potentials in quantum electrodynamics (viz-a-viz Wheeler-Feynman absorber theory).
The key insight is that quantum entanglement manipulation can effectively allow a quantum metrologist to "send back in time" information about the optimal measurement settings - information that would normally only be available after an experiment is complete. While this simulated time travel sometimes fails, when it succeeds it enables measurements that extract more information per probe than would be classically possible.
This work demonstrates a fascinating connection between quantum entanglement and apparent retrocausal effects that can generate real operational advantages forbidden by classical physics. While falling short of actual time travel, it shows how the strange properties of quantum mechanics might be harnessed in unexpected ways that challenge our usual notions of causality while potentially enabling practical improvements in quantum sensing and measurement.
Perhaps most significantly, understanding the full mechanics of what is occurring during retrocausal protocols gives insight into quantum theory itself. Since the P-CTC protocols do not violate the weak principle of causality they cannot be used for faster-than-light communication, and in that way the results remain the same as in orthodox quantum formalisms; the only difference is the interpretation of what is occurring. Positing a retrocausal mechanism removes the need for “state reduction” or a collapse of the wavefunction (describing a superposition). This offers some conceptual advantages, and perhaps greater compatibility with relativity (e.g., CTCs and ER = EPR) for a fully cogent theory of unified physics.
“Our Gedankenexperiment demonstrates that entanglement can generate operational advantages forbidden in classical chronology-respecting theories.” - Arvidsson-Shukur et al., 2024 [15].
The experiment is yet to be performed, and the latest data only comes from testing the scenario in a simulation, however this provides evidence of the feasibility of the proposed protocol, which is out of the norm to say the least; imagine telling friends and colleagues that you are working on an experiment to teleport particles back in time.
Probabilistic Instantaneous Quantum Computation
The retrocausal teleportation protocol stems from the related notion of retrodiction in quantum mechanics, which as we saw is most easily explained via a retrocausal mechanism. A highly interesting corollary that also emerges from this line of investigation—and which has significant implications for quantum computation technology—is that the principle of teleportation can be used to perform a quantum computation even before its quantum input is defined [16]. This effectively would enable instantaneous quantum computation by utilizing the entanglement state of qubits whose past state is amenable via future BSMs (Bell state measurements).
This procedure essentially implements the QT and P-CTC methodologies to receive, in a probabilistic manner, some outputs of a quantum computation instantaneously, such that if a quantum computation would normally take an arbitrarily long time, a P-CTC protocol can be utilized to obtain the exact output state instantaneously. The hypothetical procedure is outlined as follows (Figure 3):
Figure 3. a) Conventional scheme: At time t1 the engineer is given the input qubits 1 of the quantum computation (QC) in a quantum state unknown to her. She feeds them into her quantum computer and starts the computation. The computation is very time-consuming, so that the quantum computer does not terminate before the deadline at t2. b) Scheme for instantaneous quantum computation: At a time earlier than t1 the engineer has fed qubits 3, which are each maximally entangled with one qubit 2, into her quantum computer and has done the computation. At the later time t1 when the input qubits 1 are given to her the engineer performs a Bellstate measurement (BSM) on each pair of qubits 1 and 2 and projects qubits 3 onto a corresponding state. In a certain exponentially small fraction of cases the computational time is saved completely as she immediately knows that qubits 3 are projected onto the output state resulting from the correct input one. Image and image description from- Č. Brukner, J.-W. Pan, C. Simon, G. Weihs, and A. Zeilinger, “Probabilistic instantaneous quantum computation,” Phys. Rev. A, vol. 67, no. 3, p. 034304, Mar. 2003, doi:10.1103/PhysRevA.67.034304 [16].
Can quantum time travel be utilized beyond improving quantum computers, like sending people “back” in time? Some of the originators of the concept of P-CTC like Seth Lloyd caution against such notions because the retrocausal teleportation protocols currently require strong quantum entanglement, which requires stringently controlled isolation and hence there is no way that all the particles of an entire human body are going to be prepared in a strong entanglement state and enable trans-temporal activity. However, recent advancements in teleportation techniques have found that QT can be achieved in the presence of environmental interactions (noise) by utilizing multipartite hybrid entanglement of many-body systems [17]. So, don’t discount the possibility out-of-hand, because there are indications that the weak and strong principles of causality can in special circumstances be bypassed and the multiply connected entanglement network of spacetime [18] is accessible even to large quantum systems like humans.
Highlights
The retrocausal teleportation protocol represents one of the most fascinating frontiers in quantum physics, where the classical notion of cause and effect meets the bizarre quantum realm. Through careful manipulation of quantum entanglement, researchers have theoretically demonstrated that it may be possible to influence quantum states in the past, without violating the fundamental principles of relativity or causality that govern our universe.
The protocol builds upon conventional quantum teleportation techniques, which already enable the transfer of quantum states between locations using entanglement and classical communication. However, this new approach takes quantum teleportation into uncharted territory by leveraging closed timelike curves (CTCs) - theoretical constructs that emerge naturally from Einstein's field equations and quantum mechanics. While actual time travel remains firmly in the realm of science fiction, these quantum simulations of CTCs could enable practical advantages in quantum metrology and computation.
Perhaps most remarkably, the retrocausal protocol suggests that quantum entanglement can be used to effectively send information about optimal measurement settings "back in time" - information that would normally only be available after an experiment is complete. While this process is probabilistic and cannot be used for faster-than-light communication or sending macroscopic objects through time, it demonstrates how quantum mechanics continues to challenge our conventional understanding of causality and time's arrow.
The implications extend far beyond pure theoretical physics. The ability to perform quantum computations before their inputs are even defined, as demonstrated in probabilistic instantaneous quantum computation, could revolutionize quantum computing technology. Moreover, recent advances in multipartite hybrid entanglement suggest that these effects might be achievable even in noisy, real-world conditions.
As we continue to probe the boundaries between quantum mechanics and relativity, the retrocausal teleportation protocol serves as a powerful reminder that our universe is far stranger and more interconnected than classical physics would suggest. The multiply connected nature of spacetime through quantum entanglement - elegantly captured in the ER=EPR correspondence - hints at a deeper reality where past, present, and future may be more intimately linked than we ever imagined.
While the time travel of science fiction may remain far too advanced for our current understanding of spacetime, these quantum protocols are opening new windows into the nature of time itself, suggesting that causality at the quantum level is far more subtle and fascinating than our everyday experience would suggest. As we continue to develop and refine these techniques, they may not only advance our technological capabilities but also fundamentally reshape our understanding of reality's temporal fabric.
References
1. Schilpp, Paul Arthur, and Albert Einstein. Albert Einstein, Philosopher-Scientist. [1st ed.], Library of Living Philosophers, 1949. “Einsteins’s Reply to Criticisms in relation to Epistemological Problems in Atomic Physics.” Accessed: Dec. 02, 2024. [Online]. Available: https://www.marxists.org/reference/archive/einstein/works/1940s/reply.htm.
2. S. J. Olson and T. C. Ralph, “Entanglement between the Future and the Past in the Quantum Vacuum,” Phys. Rev. Lett., vol. 106, no. 11, p. 110404, Mar. 2011, doi: 10.1103/PhysRevLett.106.110404.
3. S. J. Olson and T. C. Ralph, “Extraction of timelike entanglement from the quantum vacuum,” Phys. Rev. A, vol. 85, no. 1, p. 012306, Jan. 2012, doi: 10.1103/PhysRevA.85.012306.
4. A. Higuchi, S. Iso, K. Ueda, and K. Yamamoto, “Entanglement of the vacuum between left, right, future, and past: The origin of entanglement-induced quantum radiation,” Phys. Rev. D, vol. 96, no. 8, p. 083531, Oct. 2017, doi: 10.1103/PhysRevD.96.083531.
5. J. Maldacena and L. Susskind, “Cool horizons for entangled black holes,” Fortschritte der Physik, vol. 61, no. 9, pp. 781–811, 2013, doi: 10.1002/prop.201300020.
6. Matthew S. Leifer and Matthew F. Pusey. “Is a time symmetric interpretation of quantum theory possible without retrocausality?” Proceedings of The Royal Society A. DOI: 10.1098/rspa.2016.0607 .
7. Huw Price, “Does time-symmetry imply retrocausality? How the quantum world says “Maybe”?, Studies in History and Philosophy of Science B: Studies in History and Philosophy of Modern Physics. DOI: 10.1016/j.shpsb.2011.12.003.
8. M. Ringbauer, M. A. Broome, C. R. Myers, A. G. White, and T. C. Ralph, “Experimental simulation of closed timelike curves,” Nat Commun, vol. 5, no. 1, p. 4145, Jun. 2014, doi: 10.1038/ncomms5145.
9. S. E. D’Aurelio, M. J. Bayerbach, and S. Barz, “Boosted quantum teleportation,” Jun. 07, 2024, arXiv: arXiv:2406.05182. doi: 10.48550/arXiv.2406.05182.
10. S. Pirandola, J. Eisert, C. Weedbrook, A. Furusawa, and S. L. Braunstein, “Advances in quantum teleportation,” Nature Photon, vol. 9, no. 10, pp. 641–652, Oct. 2015, doi: 10.1038/nphoton.2015.154.
11. T. Darras et al., “Teleportation-based Protocols with Hybrid Entanglement of Light,” in 2021 Conference on Lasers and Electro-Optics (CLEO), May 2021, pp. 1–2. Accessed: Dec. 03, 2024. [Online]. Available: https://ieeexplore.ieee.org/document/9571386?utm_source=chatgpt.com.
12. S. Lloyd, L. Maccone, R. Garcia-Patron, V. Giovannetti, and Y. Shikano, “Quantum mechanics of time travel through post-selected teleportation,” Phys. Rev. D, vol. 84, no. 2, p. 025007, Jul. 2011, doi: 10.1103/PhysRevD.84.025007.
13. D. T. Pegg, “Retrocausality and Quantum Measurement,” Found Phys, vol. 38, no. 7, pp. 648–658, Jul. 2008, doi: 10.1007/s10701-008-9224-2.
14. D. T. Pegg, S. M. Barnett, and J. Jeffers, “Quantum retrodiction in open systems,” Phys. Rev. A, vol.66, no. 2, p. 022106, Aug. 2002, doi: 10.1103/PhysRevA.66.022106.
15. D. R. M. Arvidsson-Shukur, A. G. McConnell, and N. Yunger Halpern, “Nonclassical Advantage in Metrology Established via Quantum Simulations of Hypothetical Closed Timelike Curves,” Phys. Rev. Lett., vol. 131, no. 15, p. 150202, Oct. 2023, doi: 10.1103/PhysRevLett.131.150202.
16. Č. Brukner, J.-W. Pan, C. Simon, G. Weihs, and A. Zeilinger, “Probabilistic instantaneous quantum computation,” Phys. Rev. A, vol. 67, no. 3, p. 034304, Mar. 2003, doi:10.1103/PhysRevA.67.034304.
17. Zhao-Di Liu, Olli Siltanen, Tom Kuusela, Rui-Heng Miao, Chen-Xi Ning, Chuan-Feng Li, Guang-Can Guo, Jyrki Piilo. Overcoming noise in quantum teleportation with multipartite hybrid entanglement. Science Advances, 2024; 10 (18) DOI: 10.1126/sciadv.adj3435.
18. N. Haramein, W. D. Brown, and A. Val Baker, “The Unified Spacememory Network: from Cosmogenesis to Consciousness,” Neuroquantology, vol. 14, no. 4, Jun. 2016, doi: 10.14704/nq.2016.14.4.961.
Study Finds that Microtubules are Effective Light Harvesters: Implications for Information Processing in Sub-Cellular Systems
Mega-network architecture of coupled chromophore qubits on Microtubule Lattice
A remarkable study on electronic energy migration in microtubules has revealed unexpected light-harvesting capabilities in these cellular structures [1]. Published in the journal ACS Central Science, the study "Electronic Energy Migration in Microtubules" by a coalition of researchers from multiple institutions—including Princeton, Stanford, Oxford, Arizona State University, the Indian Institute of Technology in Delhi, and others— have demonstrated that microtubules, cylindrical polymers of tubulin protein, can conduct electronic energy over distances of up to 6.6 nm, comparable to some photosynthetic complexes. The crystalline order of microtubules aligns light-harvesting amino acid chromophore subunits in close enough proximity to effect relatively long-range exciton energy transfer along the cytoskeletal filaments. The findings of the study demonstrated that after photoexcitation amino acid chromophores had resonant transfer of excitation energy along the microtubule comparable in efficiency to artificial light-harvesting systems, suggesting they are natural effective light harvesting macromolecular structures and can direct coherent exciton diffusion over distances much greater than what was previously presumed from first order calculations. This finding challenges previous assumptions about the quantum properties of biological systems and may have significant implications for our understanding of cellular processes, anesthetic mechanisms implicating microtubules in cognitive processes, macromolecular optoelectrical mechanics in cellular information processing, and the development of bio-inspired technologies.
What are Microtubules
Microtubules are dynamic, cylindrical filamentary structures composed of tubulin protein subunits, playing a pivotal role in maintaining cell shape, enabling intracellular transport, cellular motility, and facilitating chromosome segregation during cell division. These polymers are integral to the cytoskeleton, providing structural support and serving as tracks for motor proteins that transport cellular cargo and are therefore instrumental to internal cellular organization. Beyond their mechanical and motility functions, microtubules have been implicated in cellular signaling via multiple mechanisms [2, 3, 4, Microtubule-Actin Network Within Neuron Regulates the Precise Timing of Electrical Signals via Electromagnetic Vortices]. Their potential roles in orchestrating cellular information processing, which increasingly appears to be multitudinous—even potentially underlying cognitive processes—have become an area of focus for many researchers.
Recent studies have uncovered intriguing quantum properties within microtubules, particularly involving aromatic amino acid residues like tryptophan [5]. These residues are capable of participating in electronic energy transfer, a process that is essential for various cellular functions. Tryptophan, known for its unique fluorescence characteristics, acts as a key player in these quantum phenomena. It contributes to the formation of energy-conducting networks within the microtubule structure, facilitating long-range energy migration and potentially supporting quantum coherence. This discovery opens new avenues for exploring the intersection of quantum biology and cellular dynamics, offering insights into the fundamental processes that underpin life at the molecular level.
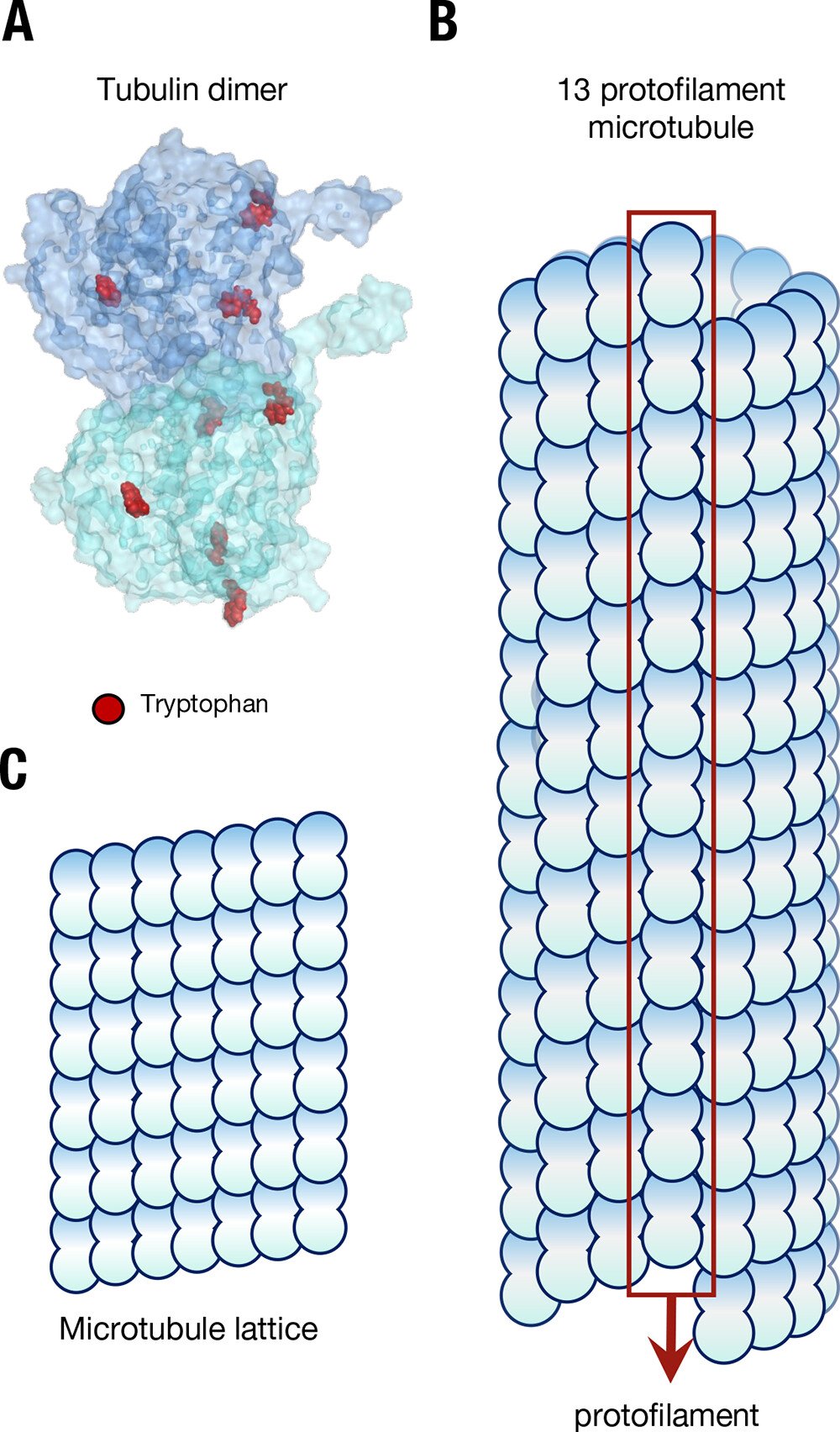
Aromatic amino acids like tryptophan are residues (i.e., subunits) of the protein tubulin (Figure 1, A) and when tubulin dimers are polymerized into helical protofilament microtubule crystalline arrays (Figure 1, B) the aromatic amino acid antennae resonators form coordinated networks, with some "mega-networks" involving up to 10,000 residues (see our article Long-range Collective Quantum Coherence in Tryptophan Mega-Networks of Biological Architectures).
Figure 1.
“The structure of microtubules forms a lattice of tubulin. (A) The tubulin dimer with tryptophan residues marked in red; the C- termini “tails” can be seen protruding from each monomer. (B) The structure of a microtubule, showing constituent arrangement of tubulin dimers, and the presence of a “seam”. (C) The repeating “lattice” of tubulin dimers in a microtubule.” Image and image description from [1] A. P. Kalra et al., “Electronic Energy Migration in Microtubules,” ACS Cent. Sci., vol. 9, no. 3, pp. 352–361, Mar. 2023, doi: 10.1021/acscentsci.2c01114.
The role of organic benzene/phenyl ‘pi electron resonance’ molecules, like tryptophan and tyrosine, as central in potential collective quantum coherent properties of microtubules has long been predicted by Stuart Hameroff, an anesthesiologist at the University of Arizona whom in collaboration with physicist and Nobel laureate Sir Roger Penrose formulated one of the first theories of consciousness involving subcellular dynamics and even quantum gravitational mechanisms [6, 7].
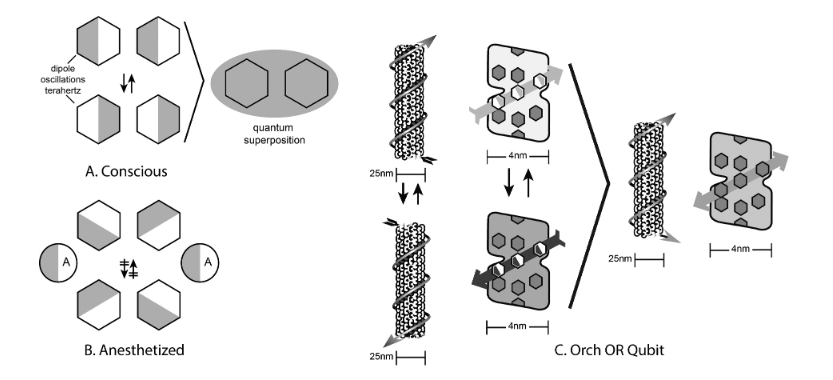
Hameroff and colleagues postulated that long-range coupling of the oscillating dipoles of aromatic amino acids residues in tubulin monomers of microtubules could process information in unique ways—such as massive parallel processing due to collective synchronization of dipole oscillators—and a proposed mechanism of orchestrated objective reduction (Orch-OR, Figure 2).
Figure 2.
“(a). Organic benzene/phenyl ‘pi electron resonance’ molecules couple, form oscillating dipoles, and quantum superposition. (b). Anesthetic gas molecules disperse dipoles, disrupt coherent oscillations, preventing consciousness. (c). The Orch OR qubit - Left: Collective dipoles oscillate in single tubulin, and along a helical microtubule pathway. Right: Quantum superposition of bothorientations in a tubulin pathway qubit.” Image and image description from [[8] S. Hameroff, “‘Orch OR’ is the most complete, and most easily falsifiable theory of consciousness,” Cognitive Neuroscience, vol. 12, no. 2, pp. 74–76, Apr. 2021, doi: 10.1080/17588928.2020.1839037.
Proposed nearly three decades ago, this theory has come to be known as the Orch-OR model, and while it has seen some criticisms during its long history, it has withstood the tests of both scrutiny and time and has recently seen wide-ranging empirical support via direct experimental observations and measurements of quantum properties of microtubules from multiple independent laboratories. One such experimental observation shedding light on the non-trivial quantum properties of microtubules is a recent study by an international research team that has found long-range electronic energy diffusion in the subcellular filaments, even confirming the disruption of such coupled electronic energy resonance transfer by anesthetic molecules, which was another prediction of Hameroff’s Orch-Or theory [9].
What was Found
The research team, led by Aarat P. Kalra and colleagues used tryptophan autofluorescence to probe energy transfer between aromatic amino acid residues in tubulin and microtubules. By studying how quencher concentration alters tryptophan autofluorescence lifetimes, they were able to quantify the extent of electronic energy diffusion in these structures.
Their results showed that aromatic amino acid residues called chromophores (chromophores are light sensitive antennas in biological macromolecules), like tryptophan and tyrosine, have robust coupling strengths over relatively long distances, with electronic energy transfer among coordinated residues occurring over approximately 6.6 nanometers for microtubules. This length of electronic energy diffusion is surprisingly high because conventionally microtubules were thought to play only structural and locomotive roles in the cell, and it is only recently that their photo-electronic processing behaviors, such as efficient electronic energy transfer, have been unequivocally identified. For comparison, chlorophyll a, a chromophore in the photosynthetic antenna complex, is specifically optimized for resonance energy transfer and absorbs photons with about 20 times the efficiency of tryptophan yet has a diffusion length of only 20-80 nm.
These findings were unexpected and challenge conventional models, such as Förster theory, which predicted electronic energy diffusion distances on the order of one nanometer for chromophore resonant transfer typical of inter-tryptophan dipole-dipole coupling, a significantly smaller distance than what was measured by Scholes et al.
Moreover, since the diffusion length of approximately 6.6 nm is on par with the size of a single tubulin dimer, which is roughly equivalent to the volume of a sphere of diameter 7.4 nm, it indicates that energy transfer between tryptophan residues could occur across a single tubulin dimer within a polymerized microtubule. As such, if a photoexcitation event occurred in a chromophore residue near an adjacent tubulin dimer, then it could be transferred along the microtubule crystalline lattice (Figure 3). Effectively resulting in coherent photon/exciton transfer, or electronic energy transfer along microtubule filaments, acting as veritable info-energy transmission filaments in subcellular macromolecular reticular networks.
Figure 3.
Schematic showing long-range energy transport along a microtubule. Image reproduced from [1].
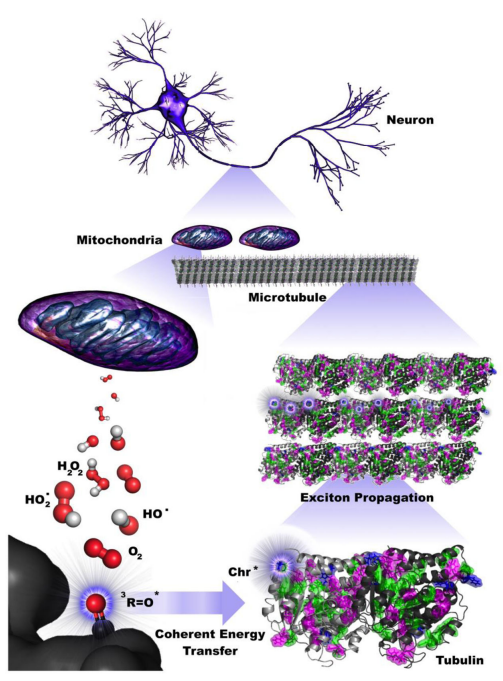
Photoexcitation events may occur continuously from reactive oxygen species production from mitochondria as described by Kurian et al., [10] (Figure 4), in which mitochondria are known to form helical filamentary networks with microtubules (Figure 5) [11].
Figure 4.
Coherent energy transfer in microtubule chromophore networks is stimulated by ultraweak photoemissions due to mitochondrial reactive oxygen species (ROS) production. Filamentous mitochondria are co-located with microtubules in the brain, suggesting that mitochondrial ROS production during respiratory activity may affect neuronal activity. Specific ROS (red and white), particularly triplet carbonyls (red and black), emit in the UV range, where aromatic networks composed of mainly tryptophan and tyrosine may be able to absorb and transfer this energy along the length of neuronal microtubules. The propagation of these excitons extends on the order of dendritic length scales and beyond, indicating that ultraweak photoemissions may be a diagnostic hallmark for neurodegenerative disease and have implications for aging processes. Image and Image description from [10] P. Kurian, T. O. Obisesan, and T. J. A. Craddock, “Oxidative Species-Induced Excitonic Transport in Tubulin Aromatic Networks: Potential Implications for Neurodegenerative Disease,” J Photochem Photobiol B, vol. 175, pp. 109–124, Oct. 2017, doi: 10.1016/j.jphotobiol.2017.08.033.
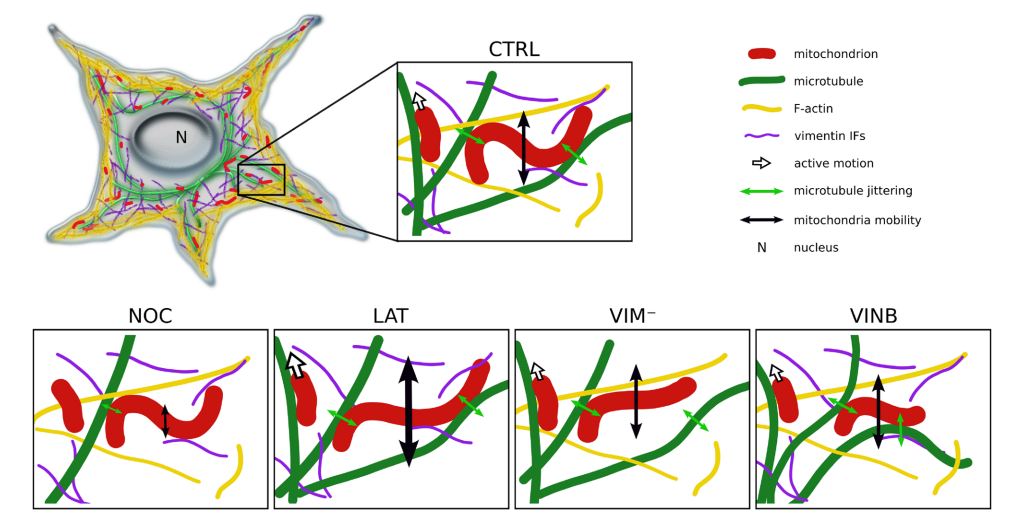
Figure 5.
Summary of the modulation of mitochondrial shape fluctuations and mobility by the cytoskeleton. Mitochondria are in close association with microtubules, being transported through them and modifying their shape as a consequence of the jittering transmitted by these filaments (green double arrows) and the interactions with F-actin and vimentin IFs, both of which would contribute to maintain mitochondria confined to microtubule network. Upon partial depolymerization of microtubules (NOC), both the mobility of the organelles (schematized with the black double arrows) and the mechanical force imposed on them decrease. Given the disruption of F-actin (LAT) and vimentin IFs (VIM−) networks, a predominance of elongated mitochondria is observed, suggesting that these filaments also modulate the organelles’ shape. F-actin depolymerization also results in increased mitochondrial mobility, suggesting that these filaments impose greater spatial confinement that restricts their motion. Perturbation of microtubule dynamics (VINB) decreases mitochondrial curvature and length compared to the control condition (All images created by A.B. Fernández Casafuz). Image and Image description from [11] A. B. Fernández Casafuz, M. C. De Rossi, and L. Bruno, “Mitochondrial cellular organization and shape fluctuations are differentially modulated by cytoskeletal networks,” Sci Rep, vol. 13, no. 1, p. 4065, Mar. 2023, doi: 10.1038/s41598-023-31121-w.
Interestingly, the researchers found that while diffusion lengths were influenced by tubulin polymerization state (free tubulin versus tubulin in the microtubule lattice), they were not significantly altered by the average number of protofilaments (13 versus 14). This suggests that the energy transfer properties are intrinsic to the tubulin structure rather than dependent on specific microtubule architectures.
How the Study was Performed
The researchers employed a multi-faceted approach to investigate electronic energy migration in microtubules. Their methodology included:
Steady-state spectroscopy: This technique was used to measure the absorption and fluorescence spectra of tubulin and microtubules under various conditions.
Time-correlated single photon counting (TCSPC): This advanced method allowed the team to measure tryptophan fluorescence lifetimes with high precision, providing crucial data on energy transfer dynamics.
Negative stain electron microscopy: This imaging technique was used to confirm the polymerization states and structures of the tubulin assemblies studied (Figure 6).
Figure 6.
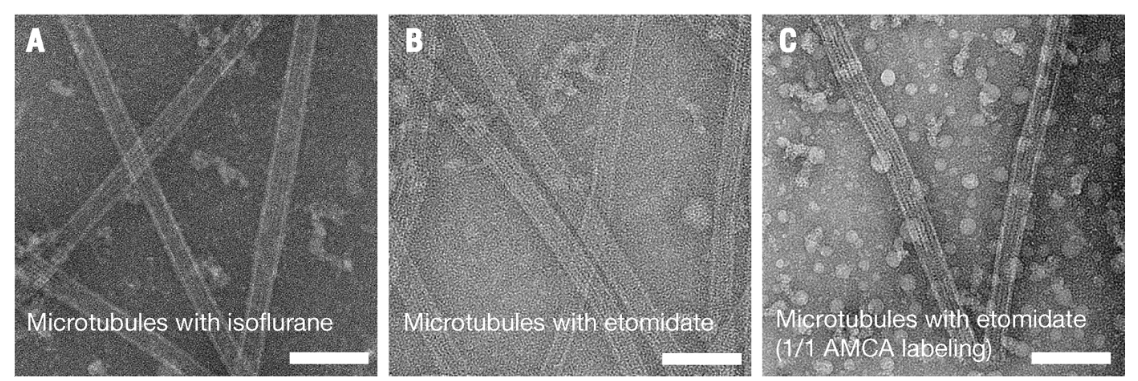
Tunneling electron microscopy validating microtubule polymerization in anesthetic containing solutions (A) isoflurane, (B) etomidate, (C) etomidate and microtubules polymerized using tubulin labeled with AMCA. Scale bars represent 100 nm. Image from [1].
In addition to these experimental methods, the team also performed computational simulations to model energy migration in microtubules. They created a computational microtubule model composed of 31 stacked tubulin rings and used it to calculate coupling strengths for energy transfer between tyrosine and tryptophan residues.
The researchers also investigated the effects of anesthetics on energy transfer in microtubules. They introduced etomidate and isoflurane into their assays and measured their impact on tryptophan fluorescence quenching.
Investigation of Anesthetic Action on Energy Transfer in Microtubules
The researchers conducted a detailed investigation into how anesthetics influence energy transfer within microtubules, since it has been a long-time theory of Hameroff and his collaborators that anesthetics work—at least in part— via inhibitory action of microtubule function, most probably via disrupting dipole-dipole coupling and hence halting resonance energy transfer and long-range collective coherence. By introducing anesthetics such as etomidate and isoflurane into their experimental assays, the research team were able to observe changes in tryptophan fluorescence quenching, a method used to assess the impact of these substances at a molecular level.
The researchers were able to empirically explore whether microtubules might facilitate quantum processes that are involved with cognitive processes and awareness. Anesthetics, known for their ability to induce unconsciousness, were found to interfere with energy transfer in microtubules, suggesting a possible link between microtubule dynamics and conscious states.
The findings propose that the disruption of energy transfer mechanisms by anesthetics could inhibit the quantum processes within microtubules necessary for consciousness.
Anesthetic drugs are effective in organisms ranging from paramecia, to plants, to primates (suggesting elements of proto-cognition are operable even in unicellular and aneural organisms) and are known to have targets in the cytoskeleton, ion channels, and mitochondria [12]. Moreover, several recent studies have implicated non-chemical quantum properties like nuclear spin and magnetic field effects on anesthetic potency [13], which again highlights the potential role of non-trivial quantum effects in microtubules that are verifiably affected by anesthetics.
The action of anesthetics on excitation energy transfer was found to alter tryptophan fluorescence lifetimes when tested via spectroscopic analysis. Introducing anesthetics, etomidate and isoflurane, into microtubule assays was shown to decrease diffusion lengths of excitation energy transmission, affect dipole-dipole interactions, reduce exciton diffusion, dampen electronic energy migration, interfere with long-range interactions, potentially inhibit dipole-based information processing, and therefore overall impacting the efficiency of electronic energy migration in microtubules. This behavior, now directly observed in experiment, supports the hypothesis [14] that long-range dipole-switching of aromatic residues for information processing is an active mechanism in the cellular cytoskeleton network.
Overall, this line of research opens new avenues for understanding how anesthetics modulate consciousness and highlights the significant role microtubules might play in cognitive functions.
Potential Insights to Glean from the Study
The findings of this study have far-reaching implications for our understanding of quantum effects in biological systems. For years, many experts believed that the biological environment was too "wet, noisy, and warm" for non-trivial quantum effects like long-range collective quantum coherence to occur. This study provides strong evidence to the contrary, demonstrating that life has indeed leveraged non-trivial quantum mechanical phenomena for its own benefit (note, there is the trivial sense in which quantum mechanics is operable in the biological system, e.g., determining electron orbital configurations and holding together molecules, non-trivial refers to QM effects apart from those that are obviously at play).
These results lend support to theories such as the Unified Spacememory Network proposed by Haramein and Brown [15], and the Orch-OR theory of Hameroff and Penrose. Both of these theories involve subcellular macromolecular assemblies like microtubules playing crucial roles in information processing and potentially in cognition and awareness.
The study also has implications for our understanding of anesthetic mechanisms. The finding that the presence of anesthetics like etomidate and isoflurane reduced exciton diffusion in microtubules is a direct indication that these optoelectrical cellular filaments and associated photon/exciton transfer dynamics are involved in information and energy processes correlated with consciousness. This observation aligns with theories that anesthetics may work by interfering with quantum coherence in neuronal microtubules.
As well, the exciton vibrational resonance energy transport is corroborative of similar studies like that by Geesink et al. in which quantum coherence and entanglement play an integral role in information processing dynamics by these subcellular structures. In a study Geesink and Schmieke found that microtubule frequencies comply with two proposed quantum wave equations of respective coherence (regulation) and decoherence (deregulation), that describe quantum entangled and disentangled states [16]. The research team also suggests that microtubules show a principle of a self-organizing-synergetic structure called a Fröhlich-Bose-Einstein state, in which the spatial coherence of the state can be described by a toroidal topology. They suggest that their study reveals an
informational quantum code with a direct relation of the eigenfrequencies of microtubules, stem cells, DNA, and proteins, that supplies information to realize biological order in life cells and substantiates a collective Fröhlich-Bose-Einstein type of behavior; further supporting the models of Tuszynski, Hameroff, Bandyopadhyay, Del Giudice and Vitiello, Katona, Pettini, Pokorny, and other prominent researchers who have posited non-trivial quantum properties of microtubules involved in cognition and awareness.
This study empirically demonstrates the long-range collective resonance of dipole oscillators in microtubules, implicating the cytoskeleton in information processing by utilizing quantum properties. By revealing the unexpected light-harvesting capabilities of microtubules, the research challenges traditional views of cellular structures and highlights their potential role in quantum coherence and information processing. This finding not only advances our understanding of the quantum realm within biology but also opens new avenues for exploring how the novel forms of matter found in the nanomachinery of life might be reverse engineered in bio-inspired technologies and perhaps even revealing something fundamental about the nature of sentience that is such a key characteristic of life and the living system.
References
[1] A. P. Kalra et al., “Electronic Energy Migration in Microtubules,” ACS Cent. Sci., vol. 9, no. 3, pp. 352–361, Mar. 2023, doi: 10.1021/acscentsci.2c01114.
[2] C. D. Velasco, R. Santarella-Mellwig, M. Schorb, L. Gao, O. Thorn-Seshold, and A. Llobet, “Microtubule depolymerization contributes to spontaneous neurotransmitter release in vitro,” Commun Biol, vol. 6, no. 1, pp. 1–15, May 2023, doi: 10.1038/s42003-023-04779-1.
[3] B. C. Gutierrez, H. F. Cantiello, and M. del R. Cantero, “The electrical properties of isolated microtubules,” Sci Rep, vol. 13, no. 1, p. 10165, Jun. 2023, doi: 10.1038/s41598-023-36801-1
[4] Singh, P., et al. "Cytoskeletal Filaments Deep Inside a Neuron Are not Silent: They Regulate the Precise Timing of Nerve Spikes Using a Pair of Vortices." Symmetry, 2021, 13(5), 821.
[5] S. Eh. Shirmovsky and D. V. Shulga, “Quantum relaxation processes in microtubule tryptophan system,” Physica A: Statistical Mechanics and its Applications, vol. 617, p. 128687, May 2023, doi: 10.1016/j.physa.2023.128687.]
[6] Hameroff, S., & Penrose, R. "Orchestrated reduction of quantum coherence in brain microtubules: A model for consciousness." Mathematics and Computers in Simulation, 1996, 40(3), 453-480.
[7] S. Hameroff, “Consciousness, Cognition and the Neuronal Cytoskeleton – A New Paradigm Needed in Neuroscience,” Front. Mol. Neurosci., vol. 15, Jun. 2022, doi: 10.3389/fnmol.2022.869935.
[8] S. Hameroff, “‘Orch OR’ is the most complete, and most easily falsifiable theory of consciousness,” Cognitive Neuroscience, vol. 12, no. 2, pp. 74–76, Apr. 2021, doi: 10.1080/17588928.2020.1839037.
[9] Kalra, A. P., Hameroff, S., Tuszynski, J., Dogariu, A., Nicolas, Sachin, & Gross, P. J. 2022, August 14. Anesthetic gas effects on quantum vibrations in microtubules – Testing the Orch OR theory of consciousness. https://osf.io/zqnjd/ Date created: 2020-04-01 Last Updated: 2022-08-14.
[10] P. Kurian, T. O. Obisesan, and T. J. A. Craddock, “Oxidative Species-Induced Excitonic Transport in Tubulin Aromatic Networks: Potential Implications for Neurodegenerative Disease,” J Photochem Photobiol B, vol. 175, pp. 109–124, Oct. 2017, doi: 10.1016/j.jphotobiol.2017.08.033.
[11] A. B. Fernández Casafuz, M. C. De Rossi, and L. Bruno, “Mitochondrial cellular organization and shape fluctuations are differentially modulated by cytoskeletal networks,” Sci Rep, vol. 13, no. 1, p. 4065, Mar. 2023, doi: 10.1038/s41598-023-31121-w
[12] M. B. Kelz and G. A. Mashour, “The Biology of General Anesthesia from Paramecium to Primate,” Current Biology, vol. 29, no. 22, pp. R1199–R1210, Nov. 2019, doi: 10.1016/j.cub.2019.09.071.
[13] H. Zadeh-Haghighi and C. Simon, “Radical pairs may play a role in microtubule reorganization,” Sci Rep, vol. 12, no. 1, p. 6109, Apr. 2022, doi: 10.1038/s41598-022-10068-4.
[14] A. P. Kalra et al., “Anesthetic gas effects on quantum vibrations in microtubules – Testing the Orch OR theory of consciousness,” Apr. 2020, Accessed: Sep. 03, 2024. [Online]. Available: https://osf.io/zqnjd/
[15] Haramein, N., Brown, W. D., & Val Baker, A. "The Unified Spacememory Network: from Cosmogenesis to Consciousness." Neuroquantology, 2016, 14(4).
[16] H. J. H. Geesink and M. Schmieke, “Organizing and Disorganizing Resonances of Microtubules, Stem Cells, and Proteins Calculated by a Quantum Equation of Coherence,” JMP, vol. 13, no. 12, pp. 1530–1580, 2022, doi: 10.4236/jmp.2022.1312095.
“Missing Law” Proposed that Describes A Universal Mechanism of Selection for Increasing Functionality in Evolving Systems
By: William Brown,
biophysicist at the International Space Federation
A recently released research article has proposed a “law of increasing functional information” with the aim of codifying the universally observed behavior of naturally evolving systems—from stars and planets to biological organisms—to increase in functional complexity over time.
To codify this behavior, it is proposed that functional information of a system will increase (i.e., the system will evolve) if many different configurations of the system undergo selection for one or more functions.
Note that “evolution” is being used in a general sense, as Darwinian evolution is regarded as specific to the biological system and requires heritable material or some form of transmissible and stable memory from one iterative variant to the next, which is conventionally not considered as operable in generic dynamic physical systems, although theories like the Unified Spacememory Network and Morphic Resonance can extend the special case of Darwinian evolution to dynamic physical systems in general as they posit a medium of transmissible memory via spacememory or a morphogenic field, respectively.
An “evolving system” is defined as a collective phenomenon of many interacting components that displays a temporal increase in diversity, distribution, and patterned behavior. As such, evolving systems are a pervasive aspect of the natural world, occurring in numerous natural contexts at many spatial and temporal scales.
While not the first such study to propose a universal mechanism to explain the near-ubiquitous observable tendency of diverse natural systems to evolve to ever increasing levels of complexity—and outstandingly, functional complexity at that—a rigorous codification approaching the level of a “natural law”—like the laws of motion, gravity, or thermodynamics—is significant. In our study The Unified Spacememory Network we proposed just such a law that generic natural systems will evolve to ever increasing levels of synergetic organization and functional complexity. In the Unified Spacememory Network approach, the ever-increasing levels of functional information of naturally evolving systems is in-part a function of the memory properties of space. In the recent study, researchers utilize a comparative analysis of equivalencies among naturally evolving systems—including but not limited to life—to identify further characteristics of this “missing law” of increasing functional complexity such as the observation that all evolving systems are composed of diverse components that can combine into configurational states that are then selected for or against based on function, and as the (often very large) configurational phase space is explored those combinations that are maximally functional will be selected for preferentially. The study also proposes mechanisms subsumed within the law of increasing functional information that account for the tendency of evolving systems to increase in diversity and generate novelty.
Universality of Evolving Systems
So certain is this that we may boldly state that it is absurd for human beings to even to attempt it, or to hope that perhaps some day another Newton might arise who would explain to us, in terms of natural laws unordered by intention, how even a mere blade of grass is produced. Kant, Critique of Judgement (1790)
The universe is replete with complex evolving systems—the universe itself can be considered an evolving system (Figure 1)—and a major endeavor of unified science is to understand and codify the underlying dynamics generating evolving systems and resulting complexification, whether spontaneous emergence in self-organizational systems or delineable underlying ordering mechanisms that verge on operational “laws of nature”. From studies such as A unifying concept for Astrobiology by E.J. Chaisson that quantitatively defines evolving systems based on energy flow, such that all ordered systems—from rocky planets and shining stars, to buzzing bees and redwood trees—can be judged empirically and uniformly by gauging the amount of energy acquired, stored and expressed by those systems [1], to Antonis Mistriotis’ universal model describing the structure and functions of living systems [2] in which evolving systems like life are identified as “far-from-the-equilibrium thermodynamic phenomenon that involves the creation of order (reduction of internal entropy) by accumulating and processing information,” there is a strong foundation within this field of investigation for understanding the physics of life and complex dynamical systems in general.
An open question within understanding the complexification of matter over time is whether there are natural laws—akin to the codification of statistical averages as laws underlying thermodynamics—that are operational in generic complex dynamical systems that can be characterized as having an asymmetric-time evolution. Chaison defines complexity as: “a state of intricacy, complication, variety or involvement, as in the interconnected parts of a system—a quality of having many interacting, different components” and notes that “particularly intriguing is the potentially dramatic rise of complexity within the past half-billion years since the end of the pre-Cambrian on Earth. Perhaps indeed resembling a modern form of Platonism, some underlying principle, a unifying law, or an ongoing process creates orders and maintains all structures in the Universe, enabling us to study all such systems on a uniform, level ground.”
Figure 1.
A stylized arrow of time highlighting the salient features of cosmic history in terms of an evolutionary process. From its supposed high-energy origins some 14 GA (left) to the here and now of the present (right) with complex evolving systems giving rise to culture, cybernetics, and AI. Labelled diagonally across the top are the major evolutionary phases that have produced, in turn, increasing amounts of order and complexity among all material systems: particulate, galactic, stellar, planetary, chemical, biological, and cultural evolution. Cosmic evolution encompasses all of these phases. Image and image description from Chaisson [1].
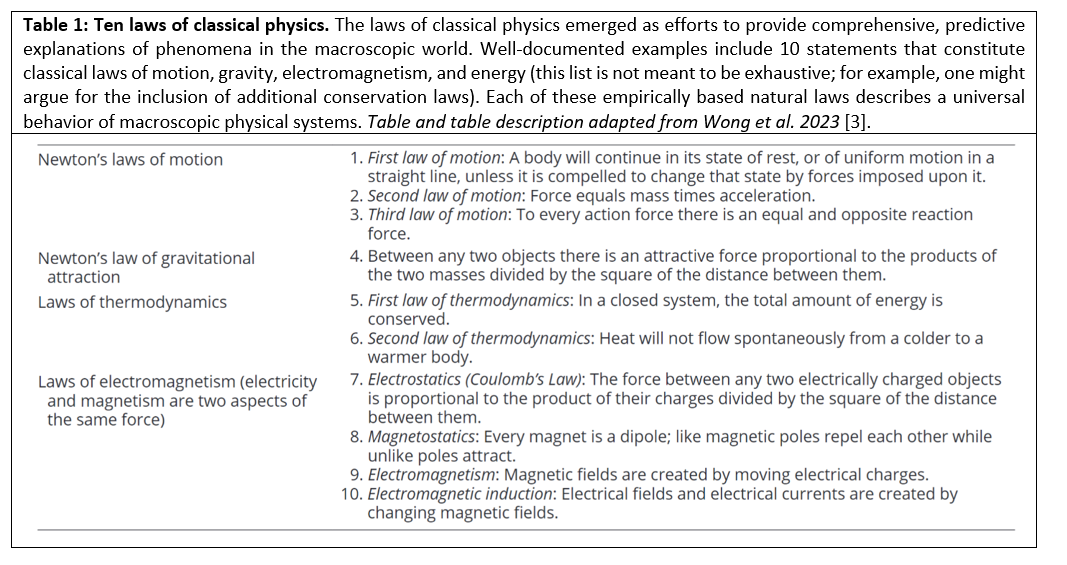
Now, there has been new advancement in this investigation into the nature of complex evolving systems as a recently release study On the Roles of Function and Selection in Evolving Systems, by Wong et al., discusses how the existing macroscopic physical laws (Table 1) do not seem to adequately describe these (complex evolving) systems [3]. Historically it has been generally accepted that there is no such equivalent universal law operational in the development and evolution of dynamic systems describing a tendency to increase in functional complexity because it is assumed that the underlying dynamics are intrinsically stochastic (randomly determined; having a random probability distribution or pattern that may be analyzed statistically but may not be predicted precisely): and therefore any general developmental or complexification process proceeds via one random accident after another with no underlying natural directionality, ordering process, or mechanism that would equate to a physical law from which, for example, a near-precise probability outcome could be made for the behavior and trajectory of any given evolving system (even though many systems, and certainly complex dynamic systems are non-deterministic).
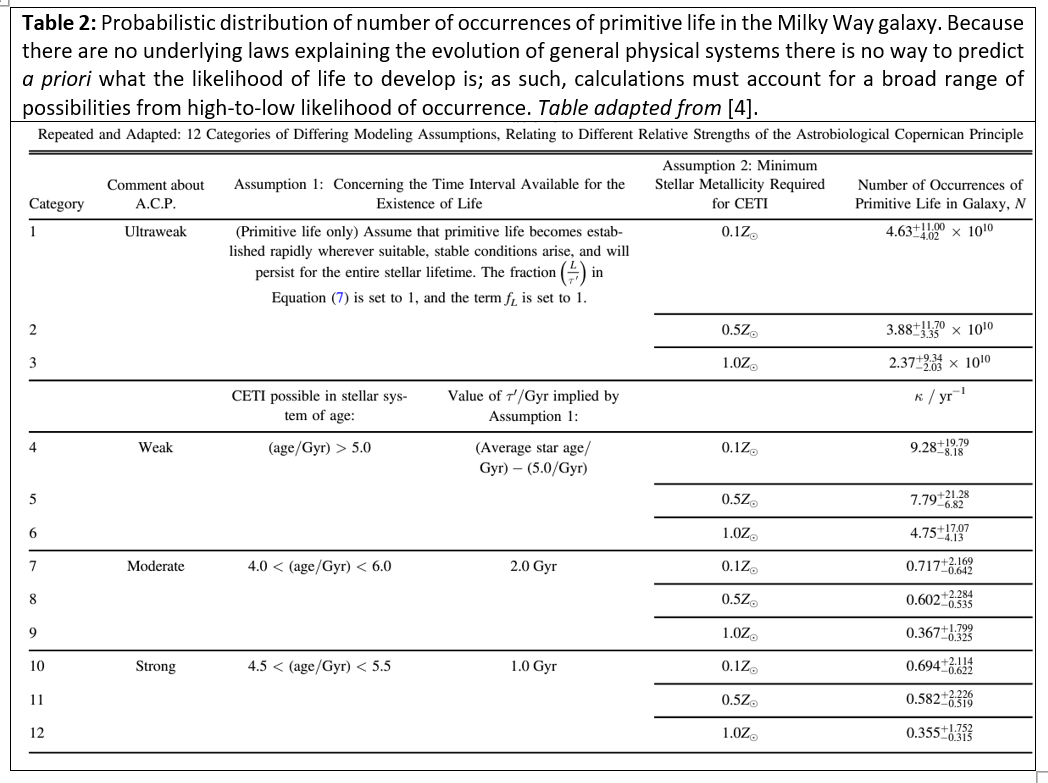
As such, in studies like The Astrobiological Copernican Weak and Strong Limits for Intelligent Life [4], by Westby and Conselice, where utilizing data to calculate the prevalence of intelligent life in the Milky Way galaxy the researchers must resort to a probabilistic analysis that takes into account a range of possibilities from a “strong scenario” with strict assumptions on the improbability of matter evolving into living organisms on any given habitable exoplanet to an “ultraweak scenario” that is more permissive in the underlying assumptions [Table 2]. So, for example, under the most permissive (ultraweak) assumptions they calculate a prevalence of approximately 4.63 X 1010 (~40 billion) number of occurrences of primitive life developing on planets in our galaxy, while under the most stringent (strong) assumptions they find that there should be at least 36 intelligent (communicating) civilizations in our galaxy, if however restrictions are loosened and calculations are made under the assumption that life has a relatively decent probability of developing on rocky planets where there is liquid water and a consistent low-entropy energy source then the number of potential intelligent civilizations in our galaxy is exponentially larger than a mere 36.
If there were known macrophysical laws delineating the behavior of evolving systems, the researchers Wesby and Conselice would not have to rely on “assumptions” for their analysis. Aside from the seemingly unscientific capitulation of attributing development of generic evolving systems—not just life—to randomness that is prevalent within conventional academia, or relying on purely emergent ordering behavior that can be spontaneously exhibited in self-organizing systems [see Kauffman, 5], this orthodox purview seems to neglect significant observables like the uniform increase in complexity and diversity of matter that is readily evident over the universe’s history and the remarkable instance of matter to organize into the superlative functionally complex system of the living organism. Indeed, the predominance of the assumption that randomness is fundamental, and emergence of order is most rationally attributed only to coincidental or serendipitous accidents leads theorists to posit that it must be extremely unlikely for something like biogenesis to occur, despite the more general observation that material systems across space and time have an inexorable tendency towards complexification and functional synergetic organization.
For example, Andrew Watson in Implications of an Anthropic Model of Evolution for Emergence of Complex Life and Intelligence [6] deduces that there is only a 2.6% chance for one of the major transitions in evolution of primordial molecular replicating systems to become living cells (Table 3), suggesting that it is highly unlikely for circumstances to permit biogenesis within timeframes similar to the known lifespan of Earth’s biosphere (supporting the “rare Earth” hypothesis). However, if we take into consideration the infodynamics operable from a guiding field, like Meijer et al’s informational quantum code [7], Sheldrake’s Morphic Resonance [8], or Chris Jeynes and Michael Parker’s Holomorphic Info-Entropy concept [9], we can make a rational inference that organizational dynamics, verging on a veritable effective entropic force or physical law of complexity, will drive systems to ever increasing functional organization and biogenesis will be a relatively likely and universal outcome wherever conditions are favorable for biological habitability.
This assumption within the orthodox approach is, however, shifting even within conventional circles. Evaluating the uniform increase in complexity and functionality of physical systems in the universe, Wong et al. have derived a physical law that they propound underlies the behavior of evolving systems, in which the functional information of a system will increase (i.e., the system will evolve) if many different configurations of the system undergo selection for one or more functions. By identification of conceptual equivalencies among disparate phenomena—a process that was foundational in developing previous laws of nature like those in Table 1—the research team purports to identify a potential “missing law”. They postulate that evolving systems—including but not limited to the living organism—are composed of diverse components that can combine into configurational states that are then selected for or against based on function. Hence, via a delineation of the fundamental sources of selection: (1) static selection, (2) dynamic persistence, and (3) novelty generation; Wong et al. have proposed a time-asymmetric law that states that the functional information of a system will increase over time when subjected to selection for function(s).
The Law of Increasing Functional Information
The laws presented in Table 1 are some of the most important statements about the fundamental behavior of physical systems that scientists have discovered and articulated to date, yet as Wong et al. point out in their recent study, one conspicuously absent statement is a law of increasing complexity. Nature is replete with examples of complex systems and a pervasive wonder of the natural world is the evolution of varied systems, including stars, minerals, atmospheres, and life (Figure 2). The study On the Roles of Function and Selection in Evolving Systems delineates at least 3 definitive attributes of evolving systems that appear to be conceptually equivalent: 1) they form from numerous components that have the potential to adopt combinatorially vast numbers of different configurations; 2) processes exist that generate numerous different configurations; and 3) configurations are preferentially selected based on function. Universal mechanisms of selection and novelty generation—outlined below—drive such systems to evolve via the exchange of information with the environment and hence the functional information and complexity of a system will increase if many different configurations of the system undergo selection for one or more functions.
Figure 2.
The history of nature from the Big Bang to the present day shown graphically in a spiral with notable events annotated. Every billion years (Ga) is represented by a 90-degree angle section of the spiral. The last 500 million years are represented in a 90-degree stretch for more detail on our recent history. Some of the events depicted are the emergence of cosmic structures (stars, galaxies, planets, clusters, and other structures), the emergence of the solar system, the Earth and the Moon, important geological events (gases in the atmosphere, great orogenies, glacial periods, etc.) and the emergence and evolution of living beings (first microbes, plants, fungi, animals, hominid species)
Origin of Selection and Function
Let’s now take a closer look at the 3 attributes identified and delineated by Wong et al. in their study; the three definitive attributes of evolving systems being: (1) static selection, identified as the principle of static persistence; (2) dynamic persistence, identified as the principle of the persistence of processes; and (3) novelty generation, a principle of selection for novelty.
Principle of static persistence (first-order selection)- configurations of matter tend to persist unless kinetically favorable avenues exist for their incorporation into more stable configurations. As described by Wong et al. persistence provides not only an enormous diversity of components but “it also provides ‘batteries of free energy’ or ‘pockets of negentropy’ throughout the universe that fuel dynamically persistent entities”.
The research team derived the first-order selection parameter of the law of increasing complexity by imagining an alternate universe that begins like our own but ultimately does not produce any systems of increasing complexity. As described, “in such a patternless world, systems smoothly march toward states of higher entropy without generating any long-lived pockets of low entropy or ‘pockets of negentropy’, for example, because of an absence of attractive forces (gravity, electrostatics)” or constants like alpha are not “fine-tuned” and stable atoms cannot even form. In our study The Unified Spacememory Netwok [10], we followed a similar thought-experiment to illustrate the mechanism of increasing synergic organization and functional complexity via the memory attribute of the multiply-connected spacetime manifold and neuromorphic ER=EPR-like connectivity architecture of the Spacememory Network (Figure 3).
Figure 3.
(A) Potential paths of the evolution of matter in the Universe (for conceptual illustration only). Arrows indicate the relative degree of probability under conventional models, with potential path 1 having the strongest degree of probability, but the lowest degree of order and complexity; potential path 2 having the lowest degree of probability, but the highest degree of ordering and complexity; and potential path 3 having a median probabilistic expectation value. (B) Postulated effect of nonlocal interactions (EPR correlations) of the ERb=EPR micro-wormhole information network on the development and evolution of atomic and molecular structures in the universe. The high density ERb=EPR micro-wormhole connections integral to complex and highly ordered molecules (pathway 2) produce a stronger interaction across the temporal dimension, as well as intramolecularly. This influences the interactivity of atoms such that there is a veritable force driving the systems to form complex associations – a negentropic effect. The trans-temporal information exchange, that appears as a memory attribute of space, is an ordering effect that drives matter in the universe to higher levels of synergistic organization and functional complexity.
Similar to our conclusion in The Unified Spacmemory Network, Wong et al. conclude that states of matter in our universe do not march smoothly to maximal entropy (pathway 1 in Figure 3), but instead there are negentropic forces that “frustrates” the dissipation of free energy “permitting the long-lived existence of disequilibria” and resulting in “pockets of negentropy” that fuel dynamically persistent entities (pathway 2 in Figure 3).
The significance of the property of certain states of matter to decrease entropy, maintain, and perpetuate far-from-equilibrium thermodynamic states as part of the complexification of evolving systems, leading to the living organism, has been pointed in previous studies like the author’s work on defining the key transition from abiotic to biotic organized matter [11]. Mistriotis has further elucidated the mechanism of negentropic action by the living systems as involving the processing of information, whereby via logic operations, like that of an electronic circuit, the internal entropy of a complex evolving system like the living organism is decreased and hence, all living systems necessarily perform logical operations similar to electronic circuits. Logic is necessary in the living system to perform the self-similar functions of decreasing entropy across the hierarchical organization of the organism, such that the similarity with the information processing of an electronic circuit is elaborated even further to draw similarities with the read-write functionality of computer memory, showing that complex evolving systems like life are processing information at a complex level [12].
Second-order selection, persistence of processes- this second postulate defines the characterization of “function” that may be attributed to a process and how functionality is ultimately selected for as opposed to process that do not contribute to the causal efficacy over internal states of a system. As described by Wong et al. “insofar as processes have causal efficacy over the internal state of a system or its external environment, they can be referred to as functions. If a function promotes the system’s persistence, it will be selected for.”
Third-order selection for novelty- the third-order selection parameter addresses a significant challenge in evolutionary theory, in which natural selection can describe the selection and preservation of adaptive phenotypes but cannot explain the de novo generation of novelty [13]. This is addressed in the new study by positing that “there exist pressures favoring systems that can open-endedly invent new functions—i.e., selection pressures for novelty generation.” Adding new functions that promote the persistence of the core functions essentially raises a dynamic system’s “kinetic barrier” against decay toward equilibrium. The new study further elaborates: “a system that can explore new portions of phase space may be able to access new sources of free energy that will help maintain the system out of equilibrium or move it even further from equilibrium. In general, in a universe that supports a vast possibility space of combinatorial richness, the discovery of new functional configurations is selected for when there are considerable numbers of functional configurations that have not yet been subjected to selection.”
Like Mike Levin’s scale-free cognition and complex organization of compound intelligences [14], in a more general sense Wong et al. point out the most complicated systems are nested networks of smaller complex systems, each persisting and helping to maintain the persistence of the whole. Importantly, in nested complex systems, ancillary functions may arise, like eddies swirling off a primary flow field.
The first, second, and third order selection-for-function parameters are proposed to account for the origins of selection and function, since the universe that we observe constantly generates certain ordered structures and patterned systems whose existence and change over time cannot adequately be explained by the hitherto identified laws of nature, such as those summarized in Table 1. These postulates lead to the formalization of a kind of law to describe the increase in system complexity through the existence of selection pressures:
Systems of many interacting agents display an increase in diversity, distribution, and /or patterned behavior when numerous configurations of the system are subject to selective pressures.
As such, there is a universal basis for selection and a quantitative formalism rooted in the transfer of information between an evolving system and its environment.
Functional Information and the Evolution of Systems
All of the natural laws in Table 1 involve a quantitative parameter such as mass, energy, force, or acceleration, and it naturally moots the question, “is there an equivalent parameter associated with evolving systems?” The latest study expounds that indeed there is, and the answer is information (measured in bits), specifically “functional information” as introduced in studies like Functional Information and the Emergence of Biocomplexity [15]. Functional information quantifies the state of a system that can adopt numerous different configurations in terms of the information necessary to achieve a specified “degree of function,” where “function” may be as general as stability relative to other states or as specific as the efficiency of a particular enzymatic reaction.
In the hierarchy of increasing complexity that characterizes the biological system, Mistriotis identifies the characteristic of Functional self-similarity, where like a fractal that repeats an elementary pattern with fixed geometric characteristics recursively to generate a complex self-similar structure, the nested hierarchical levels of the living organisms repeat functions like metabolism, growth, reproduction, and responsiveness in a self-similar pattern across each organizational domain. As such, “as fractal geometry requires an elementary pattern that acts as a seed, Functional Self-similarity implies the existence of an elementary living system” [16].
Regarding the law of increasing functional information for generic evolving systems, the functional information formalism points to an important universal characteristic:
The functional information of a system will increase (i.e., the system will evolve) if many different configurations of the system are subjected to selection for one or more functions.
As described by Wong et al. this postulate is a close parallel to the previously proposed law of increasing complexity, which states that natural selection, acting alone, tends to increase the complexity of a system [17].
Moving Towards Codifying Mechanisms of Complexification and Function as Natural Laws
The tendency for diversity and complexity to increase in physical systems has been discussed in detail in previous works, such as the proposal of Biology’s First Law by McShea and Brandon, in which they postulate a zero-force evolutionary law that states that:
In any evolutionary system in which there is variation and heredity, there is variation and heredity, there is a tendency for diversity and complexity to increase, one that is always present but may be opposed or augmented by natural selection, other forces, or constraints acting on diversity or complexity [18].
While McShea and Brandon’s proposal for a zero-force evolutionary law applies exclusively to evolutionary systems in which there is variation and heredity, and therefore is narrowly specified to living organisms (that have the hereditary capacity bestowed by the chemical memory of the DNA-RNA-protein system), the work of Wong et al expands this to generic physical systems, and in our Unified Spacememory Network postulate we describe the mechanism by which generic physical systems, or organized matter in the universe, obey a universal evolutionary force—including ever-increasing functional information, synergistic organization, complexity and diversity—via the memory properties of space.
Even within the domain of purely theoretical physics, in analysis of complexity theory, there has been proposals for concepts such as The Second Law of Quantum Complexity, by Susskind and Brown [19], where it is shown that long after a system reaches maximal entropy, its evolution is not over as at a quantum level it continues to explore combinatorial phase-space and the entanglement network of such a system will continue to complexify. As such, the quantum complexity of the system increases over time and this process far exceeds the time in which a closed system may reach equilibrium (maximal entropy state). This has important implications for black hole physics, and hence holographic and unified physics.
The Fallacy of Progress
Conventional evolutionary theory in biology is highly adverse to any proposition of an “intrinsic” tendency to increase in complexity. There are myriad examples of complex dynamical systems, however conventional theory holds that the increase in complexity is neither universal nor inevitable; Smith and Szathmary gave the example of bacteria as exemplifying this inference because many prokaryotes are probably no more complex today than their ancestors 2000 million years ago [20]. They maintain that the most we can say, in terms of the living organism, is that some lineages have become more complex in the course of time. As well, there are probably examples outside of biology of complex dynamical systems that have not exhibited a quantitative increase in their respective level of complexity, or information content, after reaching a dynamic equilibrium maximum. Smith and Szathmary further expounded key reasons why biologists tend to view any theory of an inevitable increase in complexity—amounting to a law of nature—as an erroneous inference and a fallacy, the fallacy of progress:
The notion of progress has a bad name among evolutionary biologists. Lamarck accepted the earlier idea of a ladder of nature and argued that organisms have an inherent tendency to climb the ladder. It was Lamarck’s notion of an inherent tendency, rather than his belief in the inheritance of acquired characters, that Darwin was rejecting when he said that his theory had nothing in common with Lamarck’s: he rightly saw that to explain evolution by an inherent tendency is as vacuous as to say that a man is fat because he has an inherent tendency to obesity. Today, we are unhappy with a picture of evolution that places us at the summit, and arranges all other organisms in a line behind us: what have we to be so proud about? To be fair, humans were by no means a the summit of the medieval scala naturae; there were angels and archangels above us as well as worms below.
There are, of course, more solid reasons, both empirical and theoretical, for rejecting a simple image of progress on a linear scale. Empirically, the history of life is better visualized as a branching tree than as a single ascending line. The fossil record shows that many organisms—horseshoe crabs, the coelacanth, crocodiles, for example—have undergone little change, progressive or otherwise, for hundreds of millions of years. On a shorter timescale, sibling species tell the same story. The fruit flies Drosophila melanogaster and D. simulans are hard to distinguish morphologically, but molecular data indicate that they are separated by several million years of evolution. Hence, either morphological evolution in the two species has been almost exactly parallel, which is implausible, or neither species has changed.
On the theoretical side, there is no reason why evolution by natural selection should lead to an increase in complexity, if that is what we mean by progress. At most, the theory suggests that organisms should get better, or at least no worse, at doing what they are doing right now. But an increase in immediate ‘fitness’—that is, expected number of offspring—may be achieved by losing eyes or legs as well as by gaining them. Even if an increase in fitness cannot be equated with an increase in complexity, or with progress, it might seem at first that R.A Fisher’s (1930) ‘fundamental theorem of natural selection’ at least guarantees an increase in fitness. The theorem states that the rate of increase in the mean fitness of a population is equal to the genetic variance in fitness: since variances cannot be negative, the theorem states that fitness can only increase. If so, ‘mean fitness’ in biology is an analogue of entropy in physics: it gives an arrow to time. Thus in physics the inevitable increase in entropy distinguishes past from future: if mean fitness can only increase, this defines a direction to evolution. It seems that Fisher did indeed think that his theorem could play such a role: otherwise, why ‘fundamental’? Unfortunately, the theorem holds only if the relative fitnesses of genotypes are constant, and independent of their frequencies in the population: for many traits, such constancy does not hold [ibid, 20].
So, while there may be a tendency—verging on a veritable physical law—for integrally-interacting multi-constituent dynamical systems to increase in complexity, it does not mean that every instance of such a dynamical system will necessarily increase in complexity, hence it is not absolutely necessary or universal. Some complex dynamical systems may reach an optimal information content early in their constitution, and therefore will remain relatively unchanging in an information optimum.
Unified Science- in Perspective
In the study The Autodidactic Universe [21] it is investigated whether it is possible that physical laws themselves can evolve and change. This is an interesting inquiry as it enables an evaluation of the reason why the present laws and constants of the universe are more likely than another set (known as the fine-tuning problem, which we also further discuss and clarify in our study The Unified Spacememory Network). So, for example, the coupling constants of nature (e.g., the gravitational constant G, or the fine-structure constant alpha) might turn out to be dynamical variables, and indeed in the Origin of Mass and Nature of Gravity [22] it is shown that fundamental properties like mass, the nuclear confinement or binding forces, and gravity are based on dynamical variables that are set by the conditions of decoherence of quantum vacuum fluctuations coupled with screening zero-point energy density and a resulting Planck pressure force that arises from the Planck plasma flow in black hole particles. The feedback dynamics operable in these fundamental states are quintessential information flows that characterize the organized functional change of evolving systems in the universe.
So, certainly there are mechanism underlying the complexification of our universe, and even the forces, constants, and laws themselves, which is a great thing because it means that we can come to understand these fundamental mechanisms and hence understand the nature of the universe at a deeper level. From the perspective of the Unified Spacememory Network, in which time (the 4th dimension) is holographically emergent from the memory property of 3D voxelized space—i.e., time is not fundamental—it opens an interesting consideration on just what is meant by “evolving system” if all spacetime coordinates are co-existing simultaneously. What we point out in the Spacememory Network study is that the inherently atemporal nature of the universe and its inherent nonlocal nature (as readily exemplified in quantum theory) means that there is continual crosstalk in “evolving” systems between their initial state and much later states of high functional complexity, and hence there is a trans-temporal information exchange that is the negentropic ordering force in evolution and development of physical systems. The evolutionary trajectory of a given system is just the holographic projection of its underlying neuromorphic information connectivity network, i.e., the morphogenic field and ultimately change, like time, is illusionary.
References
[1] E. J. Chaisson, “A unifying concept for astrobiology,” International Journal of Astrobiology, vol. 2, no. 2, pp. 91–101, Apr. 2003, doi: 10.1017/S1473550403001484.
[2] A. Mistriotis, “A universal model describing the structure and functions of living systems,” Communicative & Integrative Biology, vol. 14, no. 1, pp. 27–36, Jan. 2021, doi: 10.1080/19420889.2021.1887549
[3] M. L. Wong et al., “On the roles of function and selection in evolving systems,” Proc. Natl. Acad. Sci. U.S.A., vol. 120, no. 43, p. e2310223120, Oct. 2023, doi: 10.1073/pnas.2310223120.
[4] T. Westby and C. J. Conselice, “The Astrobiological Copernican Weak and Strong Limits for Intelligent Life,” ApJ, vol. 896, no. 1, p. 58, Jun. 2020, doi: 10.3847/1538-4357/ab8225.
[5] S. A. Kauffman, The Origins of Order: Self-organization and Selection in Evolution. in The Origins of Order: Self-organization and Selection in Evolution. Oxford University Press, 1993. [Online]. Available: https://books.google.com/books?id=lZcSpRJz0dgC
[6] A. J. Watson, “Implications of an Anthropic Model of Evolution for Emergence of Complex Life and Intelligence,” Astrobiology, vol. 8, no. 1, pp. 175–185, Feb. 2008, doi: 10.1089/ast.2006.0115.
[7] Meijer D K F, Wong KW, 2022. How the Universe Orchestrated the Conditions for First Life, using an Informational Quantum Code. The Concerted Action of Magnetic Monopole and Photon/Phonon Fields through a 5D Symmetry Breaking. https://www.researchgate.net/publication/357312383_How_the_Universe_Orchestrated_the_Conditions_for_First_Life_using_an_Informational_Quantum_Code_The_Concerted_Action_of_Magnetic_Monopole_and_PhotonPhonon_Fields_through_a_5D_Symmetry_Breaking
[8] Sheldrake, R. (2009). Morphic Resonance: The Nature of Formative Causation. Ukraine: Inner Traditions/Bear.
[9] M. C. Parker and C. Jeynes, “Maximum Entropy (Most Likely) Double Helical and Double Logarithmic Spiral Trajectories in Space-Time,” Sci Rep, vol. 9, no. 1, p. 10779, Dec. 2019, doi: 10.1038/s41598-019-46765-w.
[10] N. Haramein, W. D. Brown, and A. Val Baker, “The Unified Spacememory Network: from Cosmogenesis to Consciousness,” Neuroquantology, vol. 14, no. 4, Jun. 2016, doi: 10.14704/nq.2016.14.4.961.
[11] W. Brown, “Provisional Definition of the Living State: Delineation of an Empirical Criterion that Defines a System as Alive,” Qeios online journal, preprint, Jul. 2023. doi: 10.32388/V5EDGF.
[12] A. Mistriotis, “Mathematical and physical considerations indicating that the cell genome is a read-write memory,” Progress in Biophysics and Molecular Biology, vol. 178, pp. 50–56, Mar. 2023, doi: 10.1016/j.pbiomolbio.2023.01.006.
[13] A. Wagner, Arrival of the Fittest, New York City: Penguin Group, 2014.
[14] M. Levin, “The Computational Boundary of a ‘Self’: Developmental Bioelectricity Drives Multicellularity and Scale-Free Cognition,” Front. Psychol., vol. 10, p. 2688, Dec. 2019, doi: 10.3389/fpsyg.2019.02688.
[15] R. M. Hazen, P. L. Griffin, J. M. Carothers, and J. W. Szostak, “Functional information and the emergence of biocomplexity,” Proc. Natl. Acad. Sci. U.S.A., vol. 104, no. suppl_1, pp. 8574–8581, May 2007, doi: 10.1073/pnas.0701744104.
[16] Mistriotis A. Self-similarity in living systems. Amazon. com Inc.; 2018 [accessed on November 11, 2023]. Kindle e-book: https://www.amazon.com/Self- similarity-Living-Systems-Antonis-Mistriotis-ebook /dp/B07G2DVMDL
[17] D. W. McShea, Functional complexity in organisms: Parts as proxies. Biol. Philos. 15, 641–668 (2000).
[18] Daniel W. McShea and Robert N. Brandon, Biology’s First Law: The Tendency for Diversity and Complexity to Increase in Evolutionary Systems. Chicago and London: The University of Chicago Press, 2010.
[19] A. R. Brown and L. Susskind, “The Second Law of Quantum Complexity,” Phys. Rev. D, vol. 97, no. 8, p. 086015, Apr. 2018, doi: 10.1103/PhysRevD.97.086015.
[20] J. M. Smith and E. Szathmary, The Major Transitions in Evolution. OUP Oxford, 1997
[21] S. Alexander et al., “The Autodidactic Universe,” arXiv:2104.03902 [gr-qc, physics:hep-th, physics:physics, physics:quant-ph], Mar. 2021, Accessed: Apr. 16, 2021. [Online]. Available: http://arxiv.org/abs/2104.03902
[22] Nassim Haramein, Cyprien Guermonprez, & Olivier Alirol. (2023). The Origin of Mass and the Nature of Gravity. https://doi.org/10.5281/zenodo.8381115
Wormhole Counterportation & Exchange-Free Computation. The future of computation: instantaneous information processing.
In previous original RSF articles we have discussed experiments that tested qubit teleportation via a traversable micro-wormhole, and teleportation of energy utilizing the intrinsic spatial correlation (quantum entanglement) of vacuum energy density. In each case, and indeed in all quantum teleportation experiments, the “sender” and “receiver” systems must exchange information first, and this exchange of information must, necessarily, occur via a classical channel (id est, at or below the speed of light). This means that while quantum teleportation is a clever method to leverage the kind of strong spatial correlation that only occurs in quantum systems to transfer an informational state or energy from one system to another with 100% fidelity, it is not the kind of teleportation we generally think of in which something is instantaneously transferred from on location to another or reconstituted from one location to another with no intervening transit. The requirement that the receiver requires information that can only be sent via a classical communication channel means that the transference will never occur faster than the speed of light (and hence, will not violate causality or the relativity of simultaneity).
Despite the issues associated with faster-than light travel and disturbing the structural integrity and the very nature of causality, there are allowable mechanisms for effective superluminal transit, such as a traversable wormhole geometry of spacetime (an Einstein-Rosen bridge), which is an effective kind of teleportation, as entering one side of an Einstein-Rosen bridge and exiting the other would constitute a quasi-instantaneous translocation. Since this multiply-connected geometry of spacetime is allowed within the framework of general relativity, it is possible—and veritably required within unified physics / quantum gravity—that wormholes exist, and hence teleportation is possible. But why in quantum teleportation is there a requirement for a classical signal— a physical exchange of information at or below the speed of light— if the teleportation is occurring via a traversable micro-wormhole as reported in the Google Sycamore quantum computer experiment? As it turns out, upon further analysis there are some significant considerations that suggests the experiment did not actually demonstrate gravitational teleportation through a micro-wormhole. The reasons for this are detailed, but in brief: it has been suggested that the function specifying the evolution of state of the qubit system, which was highly “refined” via a machine-learning procedure, did not exhibit key features expected of gravitational teleportation via a traversable wormhole. The full critique is detailed in the report Comment on “Traversable wormhole dynamics on a quantum processor” [1].
So even when there are strong indications of gravitational interaction in a qubit teleportation experiment, such as in the Google Sycamore quantum computer experiment, there are still significant challenges to showing conclusively that quantum gravitational physics were involved. Indeed, quantum gravitational experiments in the lab [2] are no easy task, but despite the challenges, physicists are moving ahead with plans for the next gravitational teleportation experiments. The next big test of quantum gravity: a full-on teleportation where a system will be reconstituted from one location to another with no intermediary exchange of information. This is teleportation in the truest sense of the term, and in addition to elucidating via direct experimentation the nature of real-world wormholes and quantum gravity, it may lead to technological breakthroughs like ‘unhackable’ communication and exchange-free quantum computers, where output information is instantaneously accessed without necessarily going through computational processing or transmission [3].
“… a counterportation experiment demonstrating the traversability of space, by means of what is essentially a 2-qubit exchange-free quantum computer, can point to the existence in the lab of traversable wormholes.” -Hatim Salih, From Counterportation to Local Wormholes
Protocol for Direct Counterfactual Quantum Communication
The proposed experiment has the goal of full teleportation without the exchange of any physical intermediaries between “sender” and “receiver”, and, if successful, will be the strongest demonstration of traversable wormhole physics yet achieved. The experiment is detailed in the journal Quantum Science and Technology [4] and based on a protocol conceived and authored by physicist Hatim Salih [5], who is an Honorary Research Fellow at the University of Bristol’s Quantum Engineering Technology (QET) Labs, and co-founder of the start-up DotQuantum. The procedure utilizes what is called counterfactual quantum communication, or what Hatim Salih terms “counterportation” (compound term of ‘counterfactual quantum teleportation’) because although it achieves the end goal of teleportation—quasi-instantaneous disembodied translocation—unlike quantum teleportation that requires the spatial exchange of a physical signal, counterportation does so without any detectable information carriers (in that sense it is counterfactual communication).
Counterfactual communication long predates quantum mechanics, and in fact we use it all the time in the process of drawing inferences. For example, following the tautology: were A to happen, B would happen; B did not happen; therefore A did not happen—we have a clear classical counterfactual inference. Counterfactual quantum communication, however, utilizes the wave-particle duality of a signal (an information-carrying qubit) and is based on the often-counterintuitive behavior of quantum systems—like electrons, photons, atoms, or ions. One such peculiar state of a quantum system is the superposition or wavefunction, based on the wave-particle duality that can make a particle, like a qubit, appear to behave as if it is delocalized like a cloud in the field, being everywhere at once and no where specific at the same time. Interrogation (i.e., measurement) of the position of such a wavefunction can make it appear as though it has “collapsed” into a definite position, and it is often described as reduction of the wavefunction. From a unified physics purview, it is known that the wavefunction never truly collapses, it just tightens up—so to speak—but still evolves unitarily with the universal wavefunction (a nonlocal waveguide that determines the trajectories of locally isolated particles; see Bohm’s Interpretation of quantum mechanics and the de Broglie-Bohm pilot wave theory).
However, with quantum information processing, the goal is to maintain the superposition state of the information-carrying particles, or qubits, so that the wavefunction can be maximally leveraged. This can present quite a challenge when it comes to computational information processing, because conventional computation processing is based on interrogation (reading) the state of the informational bits. If this is done with a quantum state, however, the wavefunction is reduced and can no longer be utilized for quantum information processing. There is a method however, to maintain the wavefunction while still accessing the information the qubit superposition state carries—and that is via “soft measurements”, or interaction-free measurements. Counterfactual computation is just one example of an interaction-free measurement, and the protocol has been experimentally verified [6,7,8].
Figure 1. A schematic diagram of Salih et al.’s protocol for counterfactual communication, where, for every bit communicated, provably no photons have been sent to Bob. Beam splitters split a photon’s probability amplitude between the two eigenstates that correspond to the photon going in each direction; in the classical case, they split the beam intensity (and field). As interference still occurs, when Bob does not block, waves on both sides still destructively interfere, so the light never returns to Alice. However, Bob’s D3 and Alice’s D0 both detect light simultaneously. Similarly, when he blocks, light goes to his blockers and Alice’s D1 simultaneously. Therefore, in both cases, as light goes between Alice and Bob, it is not counterfactual. The only way to avoid this is to force the light to end at only one point - to postselect, with information only travelling when nothing goes between Alice and Bob. Only single photons can do this. Therefore, the only way to make the protocol counterfactual is to use these, and so make the protocol quantum. Image and image description reproduced from [12]: Hance, J.R., Ladyman, J. & Rarity, J. How Quantum is Quantum Counterfactual Communication?. Found Phys 51, 12 (2021). https://doi.org/10.1007/s10701-021-00412-5.
Chained Quantum Zeno Effect
The fact that qubits in a quantum state like the wavefunction will be irreversibly altered upon interrogation (or measurement), makes for an interesting possibility to utilize the wavefunction for quantum cryptography—since any attempt at snooping by an eavesdropper in a quantum channel will cause the qubit wavefunctions to reduce and will be immediately detectable [9]. Based on interaction-free measurements, or quantum interrogation, several quantum key distribution (QKD) protocols have been developed and successfully implemented, opening the door for a counterfactual quantum communication protocol in which no information carrying qubits are needed to physically transmit cryptographic keys between sender and receiver. The basic idea of interaction-free measurement, central to both counterfactual cryptography and counterfactual computation, makes use of experimental observation that the presence of an obstructing object (acting as a measuring device) inside an interferometer setting (which produces and guides quantum waves) destroys interference even if no particle is absorbed by the object.
This has the surprising consequence that sometimes the presence of such an object can be inferred without the object directly interacting with any (interrogating) particles. The counterfactual QKD protocol utilizes the quantum Zeno effect— which refers to the fact that repeated measurement of an evolving quantum system can inhibit its evolution, leaving it in its initial state, an effect often paraphrased as “a watched kettle never boils”. By implementing a chained version of the quantum Zeno effect, information can be directly exchanged between sender and receiver with no physical particles traveling between them, realizing direct counterfactual communication.
Counterportation Through a Quantum Wormhole
Similar to the quantum energy teleportation protocol, the counterfactual quantum wormhole teleportation protocol leverages the fact that entirely separate quantum systems can be correlated without ever having interacted via the intrinsic strong spatial correlation of vacuum-entanglement (the unified spacememory network [10,11]). This correlation at a distance can then be used to transport quantum information (qubits) from one location to another without a particle having to physically traverse the intervening space, revealing the integral multiply-connected spacetime geometry of the micro-wormhole network that connects everything.
The experiment to test the counterfactual quantum wormhole teleportation (counterportation) protocol will utilize cavity quantum electrodynamics in an optical set-up, where it can be demonstrated that communication has been achieved without detectable photons traversing the channel between two communicating parties (often referred to as Alice and Bob), no measurements are carried out between an initial quantum state and a final quantum state, and hence an underlying physical state—a traversable wormhole—is what has carried the quantum information across space. Such exchange-free communication is only explainable via the Maldacena-Susskind ERb = EPR holographic correspondence, in which quantum states like entanglement of qubits are equivalent to or a manifestation of a quantum wormhole connection through space between the two systems. The experiment proposed by Hatim will implement the construction of a universal exchange-free 2-qubit computational circuit, or gate (a CNOT gate, combined with single-qubit operations), to counterfactually transport an unknown quantum state from one or more senders to one or more receivers across space, via a local wormhole—an experiment that if successful will be a demonstration of Hatim’s counterportation protocol and primary holographic correspondence conjectures of quantum gravity.
Unified Science- In Perspective
The concept of exchange-free quantum computation / communication has significant implications for the unified physics model expounded by Haramein & Brown [10, 11], which describes an entropic force engendered from the temporal entanglement and information exchange across the multiply-connected geometry and networks of spacetime, or more aptly termed the unified spacememory network. Our work explains the ordering mechanism underlying dynamical processes such as the evolution and development of general physical systems in the universe— towards ever-increasing levels of order and organizational synergetics—leading to the emergence of living forms of matter, i.e., the living system. The same info-entropic dynamic underlying generalized evolution of organized matter and living systems is unified with the propensity of some forms of organized matter in the universe to exhibit intelligence, often and arguably arising from or correlated with the attribute called consciousness, the dynamics of which we further explain are linked via a trans-temporal information exchange generating morphic resonance, via the intrinsic wormhole connectivity circuits of the quantum vacuum, and which are integral to intelligent systems and processes via the spacememory network.
For example, regarding theories for the emergence of sentient systems, it is popular within conventional scientific theory to equate consciousness with computational processes, or more specifically with neurocomputation within the brain. However, this paradigm suffers from a fundamental misunderstanding about the nature of the biological system and a significant lack of imagination in which theorists are taking our most efficient contemporaneous means of information processing, the digital computer, and attempting to equate the biologically equivalent information processor as a kind of digital organic computer. To see how this conceptual framework may be myopic consider that our level of technological sophistication comes from about two to three thousand years of development (depending on which metric you want to use, let's say starting around the era that saw the development of the Antikythera analog computer), whereas the natural technology of the biological system has had hundreds of millions of years of refinement, so it may be a little more advanced than we are even able to recognize at our current level of understanding.
The problem with the neurocomputational paradigm (and why our current computers are not an apex information processing technology) is two-fold, (1) there is no indication that the brain is storing information digitally via binary states, hence it is not a digital computer, and (2) digital computers (our most advanced present means of information processing) will seem technologically rudimentary to future quasi-instantaneous information processing systems, like the exchange-free quantum computer (utilizing traversable wormhole teleportation). Hence, the mind and the physiological correlates of mental processing are not performing sequential digital computations to, for instance, remember past states (memory) where a network of neurons represent “on” / “off” binary values— but instead, there is an instantaneous direct accession of past states (and even potential “future” states) via the temporal entanglement of the intrinsic multiply-connected wormhole network of space. A computer based on the same principles will not so much process information, but instantaneously access the output (this can also be thought of in terms of the multiverse: in which there is access to parallel universes where “answers” have already been computed and are therefore available in universes where the actual sequential information processing has not yet physically occurred).
Another salient implication is the necessity of developing a communication technology that is not limited by the speed of light. In order for human civilization to survive far into the future it must master interstellar travel. At interstellar distances, communicating via light signals (like radio transmission) is not feasible—without even regarding the power requirements of sending a strong enough signal over light-years of distance, it is completely impractical to have a multi-year transmission time (over 8 years to send and receive a message between Earth and its closest solar system, Alpha Centauri). This is why SETI has not detected any radio transmissions—technologically advanced civilizations are not communicating via electromagnetic signals. Instead, leveraging the intrinsic spatial-entanglement of the quantum vacuum, and the multiply-connected wormhole network of spacetime, it is more likely that advanced civilizations are utilizing a form of exchange-free quantum communication, and this may be our best bet for interstellar parsec-spanning communication technology. So, the protocol and experiment proposed by Hatim Salih could not be more important! And perhaps it will lead to a path of research that one day realizes quasi-instantaneous communication and computational technology.
References
[1] B. Kobrin, T. Schuster, and N. Y. Yao, “Comment on ‘Traversable wormhole dynamics on a quantum processor’”. 15 Feb 2023, https://arxiv.org/abs/2302.07897
[2] D. Carney, P. C. E. Stamp, and J. M. Taylor, “Tabletop experiments for quantum gravity: a user’s manual,” Class. Quantum Grav., vol. 36, no. 3, p. 034001, Jan. 2019, doi: 10.1088/1361-6382/aaf9ca
[3] O. Hosten, M. T. Rakher, J. T. Barreiro, N. A. Peters, and P. G. Kwiat, “Counterfactual quantum computation through quantum interrogation,” Nature, vol. 439, no. 7079, pp. 949–952, Feb. 2006, doi: 10.1038/nature04523
[4] H. Salih, “From counterportation to local wormholes,” Quantum Sci. Technol., vol. 8, no. 2, p. 025016, Mar. 2023, doi: 10.1088/2058-9565/ac8ecd
[5] H. Salih, Z.-H. Li, M. Al-Amri, and M. S. Zubairy, “Protocol for Direct Counterfactual Quantum Communication,” Phys. Rev. Lett., vol. 110, no. 17, p. 170502, Apr. 2013, doi: 10.1103/PhysRevLett.110.170502
[6] M. Ren, G. Wu, E. Wu, and H. Zeng, “Experimental demonstration of counterfactual quantum key distribution,” Laser Phys., vol. 21, no. 4, pp. 755–760, Apr. 2011, doi: 10.1134/S1054660X11070267
[7] G. Brida, A. Cavanna, I. P. Degiovanni, M. Genovese, and P. Traina, “Experimental realization of Counterfactual Quantum Cryptography.” arXiv, Jul. 27, 2011. Accessed: Apr. 03, 2023. [Online]. Available: http://arxiv.org/abs/1107.5467
[8] Y. Liu et al., “Experimental demonstration of counterfactual quantum communication,” Phys. Rev. Lett., vol. 109, no. 3, p. 030501, Jul. 2012, doi: 10.1103/PhysRevLett.109.030501
[9] C. H. Bennett, “Quantum cryptography using any two nonorthogonal states,” Phys. Rev. Lett., vol. 68, no. 21, pp. 3121–3124, May 1992, doi: 10.1103/PhysRevLett.68.3121
[10] N. Haramein, W. D. Brown, and A. Val Baker, “The Unified Spacememory Network: from Cosmogenesis to Consciousness,” Neuroquantology, vol. 14, no. 4, Jun. 2016, doi: 10.14704/nq.2016.14.4.961
[11] W. Brown, “Unified Physics and the Entanglement Nexus of Awareness,” NeuroQuantology, vol. 17, no. 7, pp. 40–52, Jul. 2019, doi: 10.14704/nq.2019.17.7.2519
[12] Hance, J.R., Ladyman, J. & Rarity, J. How Quantum is Quantum Counterfactual Communication?. Found Phys 51, 12 (2021). https://doi.org/10.1007/s10701-021-00412-5
Study Finds Human Gene Linked to Larger Brains Arose from Non-Protein Coding (“Junk”) DNA
Researchers have discovered a key process by which new genes from non-protein coding DNA undergoes mutations to enable export from the nucleus into the cellular cytoplasm where the new gene can be translated into novel polypeptides. In the new study the researchers have shown that far from being accessories, new gene products are often integral in key phenotype characteristics, such as larger brains in human-specific de novo genes from non-protein coding DNA. But before such genes can become novel protein products, they must change to escape the nuclear localization fated for long non-coding RNA sequences: the study elucidates the mutations involved in enabling nuclear export where the new gene can access the translational machinery of the ribosome, and demonstrates via knock-out and overexpression experiments the functional role of novo genes from non-protein coding DNA in organism development, like the enlargement of the cerebral cortex in humans.
By: William Brown, scientist at the Resonance Science Foundation
The origin of novel protein-coding genes de novo was once considered so improbable that it verged on the impossible. This was a major reason why gradual evolution was favored to punctuated speciation, as it was considered that it must take millions of years for a useful gene product to emerge via the blind & random processes favored by conventional theory. However, in less than a decade, and especially in the last five years, this view has been overturned by extensive evidence of gene duplication, alternative splicing mechanisms, and pre-adaptation of non-coding DNA in diverse eukaryotic lineages to generate novel functional gene products. Rather than an extremely rare occurrence, it is now evident that there is a relatively constant trickle of proto genes released into the testing ground of natural selection. Interestingly, it has been discovered that gene-product neosynthesis can occur from sections of DNA that code for seemingly useless long RNA transcripts (what are called long non-coding RNA, or lncRNA).
The phenomenon of de novo gene birth from non-protein coding DNA is surprising, because the generic structural form of protein is amyloid, so that random polypeptides are expected to be toxic, like the amyloid plaques that characterize Alzheimer’s and neurodegenerative disease. Yet, it has become clear that useful, non-toxic gene products can indeed originate de novo from non-protein coding sequences [1], and these novel products often emerge as fully functional gene products with little-to-no intermediary acclimation phases, in an all-or-nothing type of emergence— invalidating theories of gradual evolutionary exaptation and supporting the pre-adaptation model, which proposes the existence of exaggerated gene-like characteristics in new genes and an all-or-nothing transition to functionality [2].
Although gene duplication has been reported as the predominant mechanism of the origin of new genes [3, 4], there is now an abundance of data showing that new proteins also evolve from non-protein coding DNA regions [5, 6, 7]. In contrast to gene duplication— where many of the transcription initiation, messenger RNA (mRNA) splicing / editing, and ribonucleoprotein nuclear export elements are already present— the process of de novo gene birth from long non-coding RNA sequences is surprising because many of the transcriptional and mRNA processing elements required to generate a functional gene transcript are not present. They must be generated from scratch (de novo synthesis), which requires key functional mutations to enable proper splicing and editing of the RNA sequences into valid mRNA transcripts.
To elucidate this process and further demonstrate how functional gene products are generated from non-protein coding DNA sequences, a study reported in Science [and published in Nature Ecology & Evolution, 8] has identified a human-specific gene that plays a key role in developing large complex brains that arose from long non-coding RNA. The study’s authors were able to show how the gene originated from lncRNA, showing homology between the gene and lncRNA-specific sequences, and identifying key changes in RNA splice-related sequences that enabled RNA nuclear export. Via introduction of new splicing elements, the lncRNA sequences are changed to be able to leave the nucleus where there is access to the ribosome, thus becoming functional mRNA transcripts. Perhaps the most surprising result of the study is that these newly generated (de novo) genes from long non-coding RNA sequences have biological functionality.
Possible process of pre-adaptation via cycle of gene death and gene birth. Non-genic sequences provide a constant pool of proto-genes that can be exapted for de novo gene neosynthesis.
For this latter characteristic, the research team identified 74 human/hominoid-specific novel genes, half of which emerged after the ancestral split of the human and chimpanzee lineages. Selecting one of these 74 genes that is human-specific and expressed in brain development, the research team demonstrated experimentally that knock-out or over-expression of the gene in human embryonic stem cells accelerated or delayed the neuronal maturation of cortical organoids, respectively. What’s more, when the gene was ectopically expressed in transgenic mice the test organisms developed enlarged brains with higher cortical structure like enfolding, a humanlike characteristic typical of brain morphology in the cerebral cortex.
The study therefore demonstrates how a pool of proto genes from non-protein coding DNA sequences can be rapidly exapted into functional mRNA and novel proteins that underlie key phenotypic changes in speciation— like the development of enlarged complex brains, significant in the human lineage. The pre-adaptation of the proto gene reservoir is highly intriguing and certainly raises further questions, as it suggests that the exaptation of non-protein coding DNA sequences may not be entirely serendipitous but instead that there is a natural mechanism operational to generate proto genes that can be rapidly exapted in the all-or-nothing fashion of pre-adaptation.
References
[1] McLysaght, A. & Guerzoni, D. New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Phil. Trans. R. Soc. B 370, 20140332 (2015), DOI: 10.1098/rstb.2014.0332
[2] Wilson, B. A., Foy, S. G., Neme, R. & Masel, J. Young genes are highly disordered as predicted by the preadaptation hypothesis of de novo gene birth. Nat. Ecol. Evol. 1, 0146–0146 (2017) https://doi.org/10.1038/s41559-017-0146
[3] Chen, S., Krinsky, B. H. & Long, M. New genes as drivers of phenotypic evolution. Nat. Rev. Genet. 14, 645–660 (2013)
[4] Long, M., Betran, E., Thornton, K. & Wang, W. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 4, 865–875 (2003)
[5] Li, C. Y. et al. A human-specific de novo protein-coding gene associated with human brain functions. PLoS Comput. Biol. 6, e1000734 (2010)
[6] Xie, C. et al. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet. 8, e1002942 (2012)
[7] Carvunis, A. R. et al. Proto-genes and de novo gene birth. Nature 487, 370–374 (2012)
[8] N. A. An et al., “De novo genes with a lncRNA origin encode unique human brain developmental functionality,” Nat Ecol Evol, pp. 1–15, Jan. 2023, doi: 10.1038/s41559-022-01925-6
Convergent Evolution of Retrotransposon Neuronal Function: TEs and lncRNAs in Octopus Brain Drive Sophisticated Cognitive Capabilities
Compared to humans the Octopus is in many ways alien, it is an invertebrate with the only hard part being a chitinous beak, it has eight arms where most of its neuronal tissue—or brain—is located, and in many species, it can shape-shift and change the color of its integument to match its surrounding with near perfect adaptive camouflage. However, despite the many differences, it does share one similarity with that of humans: sophisticated cognitive capabilities, including problem solving, fore-thought, and creative ingenuity.
“Among invertebrates, the nervous system of coleoids is uniquely large and complex. For example, with half a billion neurons, Octopus vulgaris has 5 times the number of a mouse (Young, 1971). Coleoids have brain lobes dedicated to learning and memory (Hochner et al., 2003) and exhibit a range of complex and plastic behaviors. Nautiloid brains are simpler, containing fewer neurons, and lack specific lobes dedicated to learning and memory (Young, 1965). The association of massive recoding with the nervous system, and the fact that it is unique to the coleoids and not observed in nautilus, hint at its relationship with the exceptional behavioral sophistication of the coleiods. This idea is reinforced by the high density of editing in transcripts that encode proteins directly involved in [neuronal] excitability. ”
Since Octopus species have a rather large evolutionary distance from humans, mammals, or even vertebrates, a study of the cellular and molecular underpinning of their sophisticated cognitive capabilities can give us insight into what specific mechanisms enable and drive intelligence in animals. Interestingly, the molecular underpinnings of neuronal plasticity and intelligence are found all the way through to the core of the cell, in the genome.
In the neurons that are most associated with learning and memory, such as those found in the hippocampus of human and mammalian brains [1], there is found some of the highest activity in any tissue of transposable elements—what are popularly referred to as mobile genetic elements or jumping genes—so much so that the genomic heterogeneity produced by transposable element-generated recombination produces genomic mosaicism in the adult brain [2].
While the exact function of the somatic cell (adult cell) nuclear recombination driven by transposons and retrotransposons is unknown, particularly in functions of learning, memory, and intelligence, it is obvious that the generation of such genomic heterogeneity will produce more dynamic neuronal plasticity—neurons with a wider array of unique phenotypes and variations that can facilitate greater adaptive information processing. In my research I have been investigating whether such somatic cell nuclear recombination may play direct roles in information processing, where memory functions are taking place at the molecular (quantum) level including DNA recombination events.
What is being discovered is that it is the non-coding regions of the genome that contribute most to what makes us unique individuals, and underpin processes involved in intelligence and cognitive capabilities. Approximately 80% of the mammalian genome is transcribed in a cell-specific manner, and the majority of that cell-specific transcription is of noncoding regions—only a small portion is transcribed into protein-coding mRNAs, and the vast majority produces numerous long noncoding RNAs (lncRNAs), which come from transposable elements. These lncRNAs are emerging as important regulators in gene expression networks by controlling nuclear architecture and transcription in the nucleus and by modulating mRNA stability, translation, and post-translational modifications in the cytoplasm [3]:
Mammalian genomes encode tens of thousands of long-noncoding RNAs (lncRNAs), which are capable of interactions with DNA, RNA and protein molecules, thereby enabling a variety of transcriptional and post transcriptional regulatory activities. Strikingly, about 40% of lncRNAs are expressed specifically in the brain with precisely regulated temporal and spatial expression patterns. In stark contrast to the highly conserved repertoire of protein-coding genes, thousands of lncRNAs have newly appeared during primate nervous system evolution with hundreds of human-specific lncRNAs. Their evolvable nature and the myriad of potential functions make lncRNAs ideal candidates for drivers of human brain evolution. The human brain displays the largest relative volume of any animal species and the most remarkable cognitive abilities. In addition to brain size, structural reorganization and adaptive changes represent crucial hallmarks of human brain evolution. lncRNAs are increasingly reported to be involved in neurodevelopmental processes suggested to underlie human brain evolution, including proliferation, neurite outgrowth and synaptogenesis, as well as in neuroplasticity. Hence, evolutionary human brain adaptations are proposed to be essentially driven by lncRNAs, which will be discussed in this review. [4] G. Zimmer-Bensch, “Emerging Roles of Long Non-Coding RNAs as Drivers of Brain Evolution,” Cells, 2019.
So, we see the integral and critical role of transposable elements, retrotransposons, and lncRNAs in evolution, development, and intelligence of animals—with striking and significant examples in the human lineage, where transposable element insertions have strongly affected human evolution [5]:
The aim of this paper is an explanation for the high speed of evolution of the human lineage, which has been exceptional compared with other animals. The high speed of evolution of human lineage brain size is recognized by comparison of fossil brain sizes... Evolution of the lineage leading to humans during the last several million years was striking. In this period the brain in our lineage tripled in mass... The function of the brain also changed rapidly but there are few useful fossils. What we know is that the result was the modern human brain, which has been called the most complex thing in the universe. We believe the brain evolution was due to natural selection and genomic variation.
Now, an international team of researchers led by Remo Sanges from SISSA of Trieste and by Graziano Fiorito from Stazione Zoologica Anton Dohrn of Naples, have conducted a comprehensive study of the Octopus’ neuronal transcriptome—sequencing the body of RNA molecules within the neurons of Octopus species, which includes retrotransposable elements and long non-coding RNAs—and have characterized a remarkable similarity with the mobile genetic landscape of humans and other mammals with high cognitive functionality [6].
Top: phylogenetic tree of cadherin genes in the California two-spot octopus (blue), Homo sapiens (red), Drosophila melanogaster (orange), Nematostella vectensis (mustard yellow), Amphimedon queenslandica (yellow), Capitella teleta (green), Lottia gigantea (teal), and Saccoglossus kowalevskii (purple). I – Type I classical cadherins; II – calsyntenins; III – octopus protocadherin expansion (168 genes); IV – human protocadherin expansion (58 genes); V – dachsous; VI – fat-like; VII – fat; VIII – CELSR; IX – Type II classical cadherins. Asterisk denotes a novel cadherin with over 80 extracellular cadherin domains found in the California two-spot octopus and Capitella teleta. Bottom: schematic of California two-spot octopus anatomy, highlighting the tissues sampled for transcriptome analysis: viscera (heart, kidney and hepatopancreas) – yellow; gonads (ova or testes) – peach; retina – orange; optic lobe (OL) – maroon; supraesophageal brain (Supra) – bright pink; subesophageal brain (Sub) – light pink; posterior salivary gland (PSG) – purple; axial nerve cord (ANC) – red; suckers – grey; skin – mottled brown; stage 15 (St15) embryo – aquamarine. Skin sampled for transcriptome analysis included the eyespot, shown in light blue. Image credit: Caroline B. Albertin et al.
The research is highly salient to understanding the molecular underpinnings of intelligence capabilities in animals because it involves a species that is evolutionarily far-removed from mammals, and yet there is a convergence of the molecular genetic mechanisms underlying neuronal plasticity and adaptive information processing. Since the octopus is a veritable alien species compared to humans, the expression and function of retrotransposons and long non-coding RNAs cannot be an extant trait that results from a shared lineage, but instead has arisen, at least to a certain degree, independently—which is a strong indication of the universality of the mechanism and its importance to cellular and molecular information processing underlying intelligence and cognition.
…we report the identification of LINE elements competent for retrotransposition in Octopus vulgaris and Octopus bimaculoides and show evidence suggesting that they might be transcribed and determine germline and somatic polymorphisms especially in the brain. Transcription and translation measured for one of these elements resulted in specific signals in neurons belonging to areas associated with behavioral plasticity. We also report the transcription of thousands of lncRNAs and the pervasive inclusion of TE fragments in the transcriptomes of both Octopus species, further testifying the crucial activity of TEs in the evolution of the octopus genomes. [6] G. Petrosino et al., “Identification of LINE retrotransposons and long non-coding RNAs expressed in the octopus brain,” BMC Biol, 2022.
The finding is a trifecta of salience, as it reveals (1) a new and deeper understanding of the critical role of non-coding DNA, which comprises more than 98% of the human genome; (2) The molecular mechanisms underlying intelligence and cognitive capabilities of animals, and (3) how intelligence and cognitive capabilities emerged evolutionarily. At least one take-away to highlight from the study is that it confronts a conventional perspective on the evolution of intelligence that it must be a very slow and gradual process. The reasoning being that something as complex and sophisticated as intelligence must take a very long time to develop. However, the strong link that is developing in our understanding between intelligence and the role of transposable genetic elements also points to how sophisticated cognitive capabilities can develop rapidly, in punctuated or even salutatory evolutionary leaps, as mobile genetic elements are also integral to accelerating evolvability and rapidly generating genomic recombination events and novel phenotypes.
A retrotransposon that I have focused much of my investigations on is the primate specific Alu retrotransposon (around 10% of the human genome is Alu elements). It has diverse roles in generating novel genes by introducing alternative splice sites and direct regulation of gene expression by inserting in gene promoter regions, and it has myriad functions in regulating the RNA transcriptome. One remarkable function is what is known as RNA editing, that enables the code of mRNA transcripts to be changed, allowing the diversification of proteomes beyond the genomic blueprint.
Cytidine and adenosine deaminases are critical RNA editors that play important functions in physiological events. a The vital role of APOBEC1 editing can be observed in the production of apolipoprotein B in the gut. The C-to-U editing at residue 2153 of hepatic Apo-B100 transforms the glutamate to a stop codon and produces a truncated protein Apo-B48 in intestinal cells [4]. b In neurons, mRNA editing of the glutamate receptor 2 (GluR2) at position 607 by ADAR2 results in an adenosine to inosine change. This transforms the CAG codon for glutamine (Q) to CIG for arginine (R) as (CGG), since ribosomes read inosine (I) as guanosine (G). This neutralizes the diffusion of divalent cations and makes the receptor impermeable to calcium [7].
This is a highly advanced genomic adaptation for rapid proteomic plasticity (generating adaptive phenotypes outside of what is included in the protein-code of the genome), and is not found in most taxa of the animal kingdom. The Alu element is specific to the majority of adenosine-to-inosine mRNA transcript modifications in the human transcriptome (at least 4.6 million modification sites identified to date). This specific retrotransposon element is not found in octopus species (or any non-primate species), however the recent study by Sanges et alia has identified a long ineterspersed element (LINE) L1 and other TEs that are potentially involved in the widespread mRNA editing observed in behaviorally sophisticated coleoid cephalopods, serving a similar function to the Alu family of retrotransposons in the human genome. Interestingly, the two class of animals where RNA editing is undoubtedly highly prevalent, with dynamic epitranscriptomic regulation, are in humans [8] and cephalopod species like the octopus [9]—adding another interesting piece to the puzzle.
References
[1] S. Bachiller, Y. Del-Pozo-Martín, and Á. M. Carrión, “L1 retrotransposition alters the hippocampal genomic landscape enabling memory formation,” Brain Behav. Immun., vol. 64, pp. 65–70, Aug. 2017, doi: 10.1016/j.bbi.2016.12.018.
[2] S. R. Richardson, S. Morell, and G. J. Faulkner, “L1 Retrotransposons and Somatic Mosaicism in the Brain,” Annual Review of Genetics, vol. 48, no. 1, pp. 1–27, 2014, doi: 10.1146/annurev-genet-120213-092412.
[3] R.-W. Yao, Y. Wang, and L.-L. Chen, “Cellular functions of long noncoding RNAs,” Nat Cell Biol, vol. 21, no. 5, pp. 542–551, May 2019, doi: 10.1038/s41556-019-0311-8.
[4] G. Zimmer-Bensch, “Emerging Roles of Long Non-Coding RNAs as Drivers of Brain Evolution,” Cells, vol. 8, no. 11, p. 1399, Nov. 2019, doi: 10.3390/cells8111399.
[5] R. J. Britten, “Transposable element insertions have strongly affected human evolution,” Proc. Natl. Acad. Sci. U.S.A., vol. 107, no. 46, pp. 19945–19948, Nov. 2010, doi: 10.1073/pnas.1014330107.
[6] G. Petrosino et al., “Identification of LINE retrotransposons and long non-coding RNAs expressed in the octopus brain,” BMC Biol, vol. 20, no. 1, p. 116, Dec. 2022, doi: 10.1186/s12915-022-01303-5.
[7] T. Christofi and A. Zaravinos, “RNA editing in the forefront of epitranscriptomics and human health,” Journal of Translational Medicine, vol. 17, no. 1, p. 319, Sep. 2019, doi: 10.1186/s12967-019-2071-4.
[8] C. Lo Giudice et al., “Quantifying RNA Editing in Deep Transcriptome Datasets,” Frontiers in Genetics, vol. 11, 2020, Accessed: Jul. 11, 2022. [Online]. Available: https://www.frontiersin.org/articles/10.3389/fgene.2020.00194
[9] N. Liscovitch-Brauer et al., “Trade-off between Transcriptome Plasticity and Genome Evolution in Cephalopods,” Cell, vol. 169, no. 2, pp. 191-202.e11, Apr. 2017, doi: 10.1016/j.cell.2017.03.025.
Article originally posted on Resonance Project Foundation
Adapted for Novoscience
Organisms change over time. In order to meet the demands of environments that inevitably produce new circumstances and challenges, species must be able to change and adapt, with each generation being more-and-more fine-tuned to the particular requirements of its ecosystem. Not only does this make good common sense, it is an observable and demonstrable fact. The scientific term given to this kind of natural change and adaptation of organisms over time is evolution. While this theory is as solid an idea as ‘Earth makes the gravitational field that causes stuff to fall’—there are still specific dynamics and mechanisms that have not been completely delineated, just like there are mechanisms underlying the physics of gravity that are still being investigated (are there gravitons? are Newtonian dynamics modified in certain regimes, producing effects of dark matter? dark gravity? emergent gravity? strong gravity? etc..).
One important aspect that has emerged in the extended synthesis of evolutionary theory is the question of evolvability. To what degree do organisms have control over the processes of change that make them responsive to their environment? Obviously, having some response mechanisms that control rates of evolution—evolvability—would be significantly beneficial to organisms who, when presented with a change in the environment, need to produce rapid novel traits in progeny to meet the new challenges. William Brown, a biophysicist with the Resonance Science Foundation, has expounded novel mechanisms of evolvability that enable organisms a degree of directed adaptation, showing that evolution is not as random or blind of a process as has been presumed. An important dynamic in the process involves what Brown calls altruistic genes, which inlcude genetic elements that are shared between organisms and which transpose within the genome, having the ability to mobilize to different locations and change genetic expression.
The ability to rapidly generate different profiles of genetic expression can allow an organism a degree of diversity and flexibility in generating phenotypes (physical traits) that are better suited to environmental conditions. Additionally, altruistic genes involve gene sharing mechanisms between organisms, one proposal of which involves direct transfer via membrane-bound vesicles – which would explain the origin of viruses. Obviously, such a theory would mean that not all virus-like transfers are necessarily detrimental or pathogenic, a postulation that has seen recent observational support with the gene sharing (horizontal gene transfer) among Archea using membrane-bound exchange. Note that viruses, as well as transposable genes (which are endogenous retroviral elements) are mostly only identified when they cause illnesses—which raises the question of whether viral-mediated genetic exchange pathways that appear neutral or even beneficial would be identified. It took a great deal of work and controversy to show that transposable genetic elements undoubtedly can confer benefit via increasing epigenetic diversity and alternative gene expression.
The picture that begins to emerge is that there are a number of existing mechanisms that allow a degree of directed adaptation in which organisms can, to a certain extent, control their own evolvability. A recent study reported from NASA’s Astrobiology Institute at the University of Illinois at Urbana-Champaign, headed by physicist Nigel Goldenfeld, has shown that transposon activity increases in a population of microbes in response to environmental stress—enabling a mechanism by which the microorganisms can rapidly adapt via increased genetic and phenotypic diversity.
“Our work shows that the environment does affect the rate at which transposons become active, and subsequently jump into the genome and modify it,” Goldenfeld said. “Thus, the implication is that the environment does change the evolution rate. What our work does not answer at this point is whether the transposon activity suppresses genes that are bad in the particular environment of the cell. It just says that the rate of evolution goes up in response to environmental stress.
“This conclusion,” he added, “was already known through other studies, for certain types of mutation, so is not in itself a complete reversal of the current dogma. We hope that future work will try to measure whether or not the genome instabilities that we can measure are adaptive.”
Article: A Simple Bacterium Reveals How Stress Drives Evolution
Study Reveals Indications of Environmental Sensing by Genetic Apparatus Driving Non-random Mutation for Directional Adaptation. By: William Brown, Biophysicist at the Resonance Science Foundation
In our study The Unified Spacememory Network: from cosmogenesis to consciousness, we described how quantum information processing pathways in the molecular genetic system of the biological organism order permutations in a non-random fashion that result in natural directional adaptation and evolution. Information exchanges involving quantum mechanisms at the molecular level of the biological system with the environment and the entanglement nexus of the Spacememory morphogenic field give a kind of natural intelligence to the evolvability of organisms, allowing for meaningful adaptations to occur at an accelerated rate beyond what would be possible under purely random genetic mutations. As the evolutionary biologist Andreas Wagner states, “natural selection can preserve innovations, but it cannot create them… nature’s many innovations, some uncannily perfect, call for natural principles that accelerate life’s ability to innovate” [1].
Such directional gene adaptation had already been observed in experimentations with gene knockout-constructs with the bacteria species E. coli—in which, for example, a gene vital for the metabolism of the sugar lactose is knocked-out or made dysfunctional by introducing a small change in the nucleotide code, when placed in a lactose rich medium, bacteria are observed to “re-activate” the gene, regaining functionality by a purposeful base-change, in a timespan that is accelerated beyond what would occur if changes in the gene were occurring purely randomly [2].
Often a single nucleotide substitution, in this example changing an Adenine residue to a Guanine nucleotide— what is called a single nucelotide polymorphism (SNP) — results in an altered gene product, a protein with a novel functionality, or a dysfunctional protein in the case of disease pathology
It is almost as if the organisms had some environmental sensing apparatus of the genetic machinery, enabling non-random directional mutation, and demonstrated that selective pressures where not the only factor in determining variational phenotype outcomes, recalling Andreas Wagner’s insight that natural selection is not the only force at play and there must be other natural principles involved that are accelerating the living systems ability to generate innovations via directional adaptations.
Now, in a study conducted through the University of Haifa in Israel and University of Ghana, a team of researchers have found an accelerated rate of an adenine to thymine nucleotide substitution in the human hemoglobin subunit beta (HBB) gene, what is referred to as the hemoglobin S mutation (HbS mutation), which results in substantial protection against severe malaria in heterozygotes (and sickle cell anemia in homozygotes) [3]. The researchers point out that malaria has been the strongest known agent of selection in humans in recent history, as it has been a leading cause of human morbidity and mortality— often causing more than a million deaths per year in the recent past, making this study especially salient for understanding natural selection at the molecular genetic level and possible intrinsic mechanisms of adaptive change.
A single nucleotide substitution results in a single nucleotide variant (SNV), generating an allele, or variant genetic code. Individuals that have the same SNV from their mother and father are homozygous for that gene loci, whereas SNV sites that are different in the maternal and paternal allele are heterozygous.
The study compared origination rates of target mutations at target base positions in a 6-bp region spanning 3 codons in the HBB gene between genome samples from European and African populations, to assess if environmental pressure from increased rates of malaria infection has any outcome on the rate of de novo (occurring for the first time) mutation in the critical HbS subunit. The study found that the de novo HbS mutation was greatly accelerated—occurred with greater frequency—in the genome samples from African populations, where generation of the allele (gene variant) has much greater adaptive significance than in locales where malaria is not endemic.
"The results show that the HbS mutation is not generated at random but instead originates preferentially in the gene and in the population where it is of adaptive significance,” said Prof. Livnat [of the University of Haifa]. Unlike other findings on mutation origination, this mutation-specific response to a specific environmental pressure cannot be explained by traditional theories. “We hypothesize that evolution is influenced by two sources of information: external information that is natural selection, and internal information that is accumulated in the genome through the generations and impacts the origination of mutations,” said Livnat.
J. Shavit, “Groundbreaking study challenges evolutionary theory, says genetic mutations aren’t always random,” Brighter Side News, Feb. 01, 2022. (accessed Feb. 22, 2022).
The study is significant for molecular genetics in that it is one of the first to measure mutation rates at a specific base pair position within a single gene—because of technological limitations measurement of mutations rates had previously been limited to averages of the entire genome or across the entire stretch of a gene, limiting the resolution of variational rates to large sections of the genome and all but excluding point mutations or entire gene subunits.
Correspondence of HbS allele frequency and incidence of Malaria. The HbS adaptive mutation occurs with a much higher frequency in locations where malaria is endemic. While this allelic distribution would be expected from selective pressures of the environment (natural selection), the researchers in the latest study found that such high frequency occurrence is not only the result of selection pressure, but an internal mechanism of increased de novo mutation in the gene generating the allele variant.
The novel methodology developed by the researchers that enables identifying and counting ultra-rare genetic variants of choice in extremely narrow regions of interest, will allow similar such studies of de novo mutation rates in important gene adaptations that involve single nucleotide substitutions—many such variants that have been exemplars in the study of adaptation by random mutation and natural selection (the conventional model of evolution). As well, the high-resolution knowledge of mutation rates that this novel method enables will allow for detailed studies of recurrent genetic diseases and cancer, allowing for a better understanding of the genetic pathways of these afflictions, and possible mechanisms to mitigate their deleterious occurrence.
The research team states that they do not yet know what mechanisms could be at play to generate increased de novo mutations in this target site that confers adaptive benefits—it is almost as if the genomes of these populations are carrying some kind of memory of the environmental pressures present that enables accelerated adaptive response in target loci (gene sites)—but further high-resolution studies of single nucleotide polymorphism rates across these and other loci and organisms may uncover the molecular mechanisms that are involved.
Certainly, the study is an empirical validation of the postulate we discussed in the Unified Spacememory Network of directional non-random adaptation and evolution, and whatever the genetic mechanisms that may be at play in generating the non-random responses of the gene observed in the study, it may very well be possible that the quantum information pathways we have described are an integral component of the sensing and response apparatus.
References
[1] Wagner A. Arrival of the Fittest: Solving Evolution’s Greatest Puzzle. Penguin Group: NY, New York City, 2014.
[2] M. Pigliucci, G.B. Muller, Evolution the Extended Synthesis, pg. 33 – 44, Chance Variation Redux, 2010. The MIT Press, Cambridge, Massachusetts; London, England.
[3] D. Melamed et al., “De novo mutation rates at the single-mutation resolution in a human HBB gene-region associated with adaptation and genetic disease,” p. 35.
Novoscience Immortalization
How to cure cancer and stop aging
The following is a discourse on a potential method to augment the cellular system of the body to have a built-in molecular receptor-communication system. The method is not for the technophobic, or those who have a religious or ethical aversion to augmenting the human body through molecular engineering.
The problem with fighting cancer or aging-associated cellular senescence is that there are around 200 trillion cells comprising the human body with thousands of different cell types. Targeting cells for introduction of novel genes or even targeted molecular therapeutics is a significant challenge; for instance targeting a neoplasm with cytotoxic chemical agents is often limited in specificity and aside from some developing advanced techniques nearly all tissues of the body are poisoned. Moreover the immortalized “stem cell” at the heart of the neoplasm often evades chemotherapeutic destruction and while the cancerous cells around it are destroyed, the tumorigenic stem cell can often go on to form a new tumor later.
As well, if it is desired to target stem cells in the body to introduce DNA repair genes or activate and extend the telomeres, it is not currently possible to selectively and efficiently target the stem cells for such cellular rejuvenation therapy. A targeted delivery methodology for both of the examples given (cancer elimination and stem cell rejuvenation), requires developing gene constructs or therapeutic agents that (1) recognize specific cell-surface markers to the desired cell type; (2) bind those cell-surface receptors; and (3) are imported into the cell and integrated into the appropriate subcellular system.
A potential way to overcome this challenge is to pre-emptively load the stem cell pool of the body with an engineered surface-receptor and integration system. This would require genetic engineering of the totipotent embryonic stem cells (Figure 1). By targeting the totipotent stem cells before gastrulation all stem cells of the body will have the surface-receptor and integration system.
Figure 1: totipotent embryonic stem cell goes on to form pluripotent embryonic stem cells that comprise the three primary germ layers of the early embryo: the ectoderm (forms the skin and nervous system), mesoderm (develops into muscle, bone, and organs), and endoderm (will form the lining of the gut, liver, and the lungs).
The surface-receptor and integration complex is a multi-component system, such that the particular isoform of cell surface-receptor that is expressed on the stem cell is linked to gene expression of the particular cell type. So for instance, expression of the brain-derived neurotrophic factor (BDNF) gene will activate the designed gene construct to express a cell surface-receptor specific to neuroblasts. When the system detects abnormalities in expression of P21, a particular cancer-targeting cell surface-receptor is expressed.
Through this system, stem cells in the adult can be specifically targeted for cellular rejuvenation, where delivery of DNA repair enzymes or even gene inserts can be directed to specific stem-cell tissue types to revert the stem cell pool back to a youthful state. Or, when a neoplasm develops drugs can be targeted directly to the cancerous cells, destroying them. These therapeutics could be administered on a monthly basis, keeping the body clear of cancer and age-associated deterioration, or given in a comprehensive fashion periodically for routine cellular rejuvenation.
If the stem cells of the body are maintained indefinitely in a healthy and youthful state, and cancers are eliminated nearly as soon as they arise, there is no reason the body could not be kept in a healthy and youthful state indefinitely: this is the novoscience immortalization methodology.